Problem Description
The twenty-first century is a biology-technology developing century. We know that a gene is made of DNA. The nucleotide bases from which DNA is built are A(adenine), C(cytosine), G(guanine), and T(thymine). Finding the longest
common subsequence between DNA/Protein sequences is one of the basic problems in modern computational molecular biology. But this problem is a little different. Given several DNA sequences, you are asked to make a shortest sequence from them so that each of
the given sequence is the subsequence of it.
For example, given "ACGT","ATGC","CGTT" and "CAGT", you can make a sequence in the following way. It is the shortest but may be not the only one.
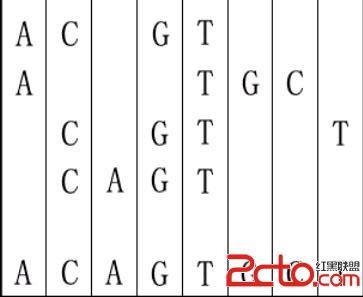
1 4 ACGT ATGC CGTT CAGT
8
題意就是給出N個DNA序列,要求出一個包含這n個序列的最短序列是多長
#include#include #include #include using namespace std; int n,deep; char c[10] = "ACGT"; struct node { char s[10]; int len; } a[10]; int pos[10];//記錄第i個序列正在使用第幾個位置 int get_h() { int ans = 0; for(int i = 1; i<=n; i++) ans = max(ans,a[i].len-pos[i]);//找出在當前情況下最長的未被匹配的長度圍毆估測長度 return ans; } int dfs(int step) { if(step+get_h()>deep)//當前長度+估測的長度比deep還大的話,也就沒有繼續往下搜索的必要了 return 0; if(!get_h()) return 1; int i,j; int tem[10]; for(i = 0; i<4; i++) { int flag = 0; for(j = 1; j<=n; j++) tem[j] = pos[j];//先將pos保存起來 for(j = 1; j<=n; j++) { if(a[j].s[pos[j]] == c[i])//當前這位符合,則該串的位置往後移一位 { flag = 1; pos[j]++; } } if(flag)//有符合的,則往下搜索 { if(dfs(step+1)) return 1; for(j = 1; j<=n; j++)//還原 pos[j] = tem[j]; } } return 0; } int main() { int t,i,j,maxn; cin >> t; while(t--) { cin>>n; maxn = 0; for(i = 1; i<=n; i++) { cin>>a[i].s; a[i].len = strlen(a[i].s); maxn = max(maxn,a[i].len); pos[i] = 0; } deep = maxn; while(1) { if(dfs(0))break; deep++; } cout << deep << endl; } return 0; }