前些天發現了一個巨牛的人工智能學習電子書,通俗易懂,風趣幽默,無廣告,忍不住分享一下給大家.(點擊跳轉人工智能學習資料)
說明:Data attachments and if requiredpythonPlease move to the WeChat public account for the source file“創享日記”,Contact the author for a fee!
一、題目
一、題目
根據附件cell.jipg,使用opencv庫或者PILThe library calculates the area ratio of the nucleus to the cytoplasm.
二、題目分析
This question is when identifying cells,To identify the nucleus and cell membrane separately.when doing this,First you need to convert the image to grayscale.Next, the grayscale image needs to be binarized,通過thresholdThe method passes in a grayscale image and a threshold,We get the nuclear threshold based on the color of the nucleus and cell membrane131,The cytoplasmic threshold is220,Then the third parameter passes in the maximum threshold255,第四個參數為THRESH_BINARY,Indicates the binarization threshold.
Then according to the size of the kernel,Pixels near the border are discarded.kernel =np.ones((2,2),np.uint8),在這裡我們使用2*2的內核,它包含了所有的1.Then pass in the image through the open operation,kernel, etc. to calculate.接著通過cannyfunction for boundary detection,Find out the contour and extract the border.通過findContoursPass in the detected edge.Finally, the border area is stored in two lists,Get the index box corresponding to the largest area,The nucleocytoplasmic ratio is obtained by dividing the calculated nuclear and cytoplasmic sizes
三、代碼
import cv2
import numpy as np
img = cv2.imread("cell.jpg")
# 轉為灰度圖像
gray = cv2.cvtColor(img, cv2.COLOR_BGR2GRAY)
cv2.imshow("gary", gray)
# 二值化
thresh1, Cellmask = cv2.threshold(gray, 220, 255, cv2.THRESH_BINARY)
thresh2, Coremask = cv2.threshold(gray, 131, 255, cv2.THRESH_BINARY)
# 開運算
kernel = np.ones((2, 2), np.uint8)
Cellmask = cv2.morphologyEx(Cellmask, cv2.MORPH_OPEN, kernel)
Coremask = cv2.morphologyEx(Coremask, cv2.MORPH_OPEN, kernel)
# 邊緣檢測
cannyCell = cv2.Canny(Cellmask, 40, 200)
cannyCore = cv2.Canny(Coremask, 40, 200)
# border extraction
Cell, Celldata = cv2.findContours(cannyCell, mode=cv2.RETR_EXTERNAL, method=cv2.CHAIN_APPROX_NONE)
Core, Coredata = cv2.findContours(cannyCore, mode=cv2.RETR_EXTERNAL, method=cv2.CHAIN_APPROX_NONE)
# Stores the border area
CellS = []
CoreS = []
for i in range(len(Cell)):
CellS.append(cv2.contourArea(Cell[i]))
for i in range(len(Core)):
CoreS.append(cv2.contourArea(Core[i]))
# Get the bounding box index corresponding to the largest area
Maxcell = CellS.index(max(CellS))
Maxcore = CoreS.index(max(CoreS))
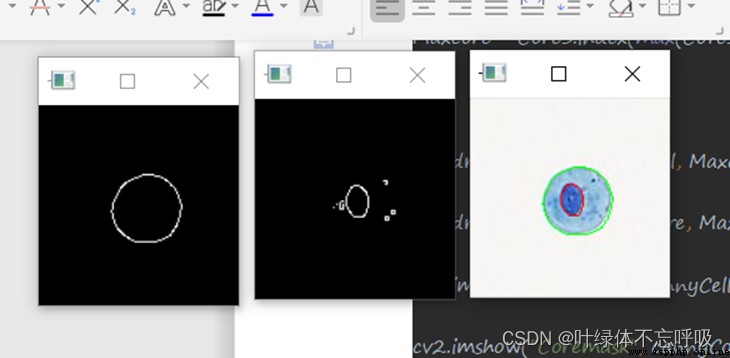
cv2.drawContours(img, Cell, Maxcell, (0, 255, 0), 1)
cv2.drawContours(img, Core, Maxcore, (0, 0, 255), 1)
cv2.imshow("Cellmask", cannyCell)
cv2.imshow("Coremask", cannyCore)
cv2.imshow("img", img)
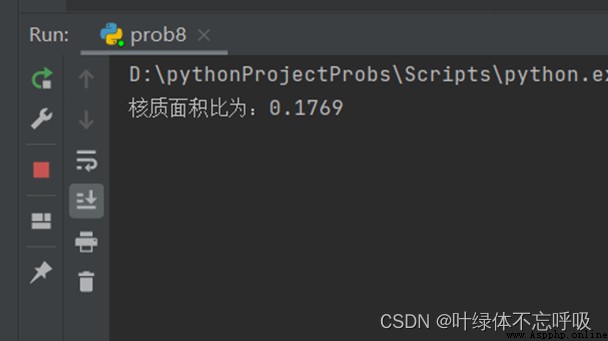
print("The nucleocytoplasmic area ratio is :{:.3f}".format(max(CoreS) / (max(CellS)-max(CoreS))))
cv2.waitKey()
四、實驗結果