Preface
CSV And text files
1 Argument parsing
1.1 Basics
1.2 Column 、 Indexes 、 name
1.3 General resolution configuration
1.4 NA And missing data processing
1.5 Date time processing
1.6 iteration
1.7 quote 、 Compression and file format
1.8 Error handling
2. Specify the type of data column
PrefaceWe introduced pandas The basic syntax operation of , Let's start with pandas Data read and write operation of .
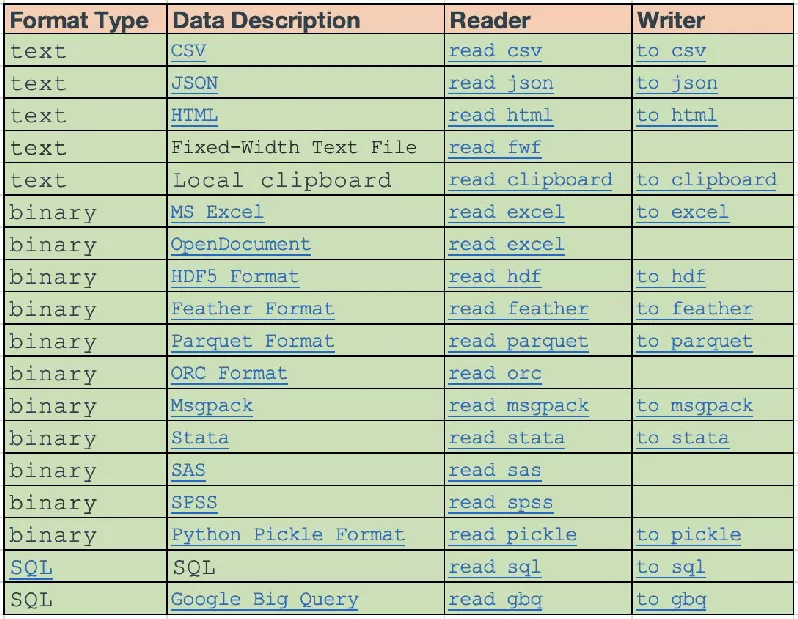
pandas Of IO API It's a group of top reader function , such as pandas.read_csv(), Will return a pandas object .
And the corresponding writer Functions are object methods , Such as DataFrame.to_csv().
All the... Are listed below reader and writer function
Be careful : We'll use that later StringIO, Please make sure to import
# python3from io import StringIO# python2from StringIO import StringIOCSV And text files The main function for reading text files is read_csv()
1 Argument parsingread_csv() Accept the following common parameters :
1.1 Basicsfilepath_or_buffer: Variable
It can be a file path 、 file URL Or any with read() Object of function
sep: str, Default ,, about read_table yes \t
File separator , If set to None, be C The engine cannot automatically detect delimiters , and Python The engine can automatically detect the separator through the built-in sniffer tool .
Besides , If the set character length is greater than 1, It's not '\s+', Then the string will be parsed into regular expressions , And mandatory use Python Parsing engine .
for example '\\r\\t', But regular expressions tend to ignore reference data in text .
delimiter: str, The default is None
sep Alternative parameters for , Consistent function
1.2 Column 、 Indexes 、 nameheader: int or list, The default is 'infer'
The line number used as the column name , The default behavior is to infer column names :
If not specified names Parameters behave like header=0, That is, infer from the first line read .
If set names, Behavior and header=None identical .
It can also be for header Settings list , Represents a multi-level column name . Such as [0,1,3], Unspecified rows ( Here is 2) Will be skipped , If skip_blank_lines=True, Blank lines and comment lines will be skipped . therefore header=0 Does not represent the first line of the file
names: array-like, The default is None
List of column names to be set , If the file does not contain a title line , Should be passed explicitly header=None, And duplicate values are not allowed in this list .
index_col: int, str, sequence of int/str, False, The default is None
Used as a DataFrame Index column of , It can be given in the form of a string name or a column index . If a list is specified , Then use MultiIndex
Be careful :index_col=False Can be used to force pandas Do not use the first column as an index . for example , When your file is an error file with a delimiter at the end of each line .
usecols: List or function , The default is None
Only the specified columns are read . If it's a list , Then all elements must be positions ( That is, the integer index in the file column ) Or a string , These strings must be the same as names The column names provided by the parameter or inferred from the document header row correspond to .
The order in the list is ignored , namely usecols=[0, 1] Equivalent to [1, 0]
If it is a callable function , Will be calculated based on the column name , The return callable function evaluates to True The name of
In [1]: import pandas as pdIn [2]: from io import StringIOIn [3]: data = "col1,col2,col3\na,b,1\na,b,2\nc,d,3"In [4]: pd.read_csv(StringIO(data))Out[4]: col1 col2 col30 a b 11 a b 22 c d 3In [5]: pd.read_csv(StringIO(data), usecols=lambda x: x.upper() in ["COL1", "COL3"])Out[5]: col1 col30 a 11 a 22 c 3Using this parameter can greatly speed up parsing time and reduce memory usage
squeeze: boolean, The default is False
If the parsed data contains only one column , So go back to one Series
prefix: str, The default is None
When there is no title , Prefix added to automatically generated column numbers , for example 'X' Express X0, X1...
mangle_dupe_cols: boolean, The default is True
Duplicate columns will be specified as 'X','X.1'…'X.N', instead of 'X'... . If there is a duplicate name in the column , Pass on False Will cause the data to be covered
1.3 General resolution configurationdtype: Type name or type Dictionary (column -> type), The default is None
The data type of the data or column . for example .
{'a':np.float64,'b':np.int32}
engine: {'c', 'python'}
The parser engine to use .C The engine is faster , and Python The engine is now more fully functional
converters: dict, The default is None
A function dictionary for converting values in certain columns . Keys can be integers , It can also be listed
true_values: list, The default is None
The data value is parsed as True
false_values: list, The default is None
The data value is parsed as False
skipinitialspace: boolean, The default is False
Skip spaces after delimiters
skiprows: Integer or integer list , The default is None
The line number to skip at the beginning of the file ( The index for 0) Or the number of lines to skip
If you can call a function , Then apply the function to the index , If you return True, Then the line should be skipped , Otherwise return to False
In [6]: data = "col1,col2,col3\na,b,1\na,b,2\nc,d,3"In [7]: pd.read_csv(StringIO(data))Out[7]: col1 col2 col30 a b 11 a b 22 c d 3In [8]: pd.read_csv(StringIO(data), skiprows=lambda x: x % 2 != 0)Out[8]: col1 col2 col30 a b 2skipfooter: int, The default is 0
Need to skip the number of lines at the end of the file ( I won't support it C engine )
nrows: int, The default is None
Number of file lines to read , Useful for reading large files
memory_map: boolean, The default is False
If filepath_or_buffer Parameter specifies the file path , The file object is mapped directly to memory , Then access the data directly from there . Use this option to improve performance , Because there's no more I/O expenses
1.4 NA And missing data processingna_values: scalar, str, list-like, dict, The default is None
Need to be converted to NA String of values
keep_default_na: boolean, The default is True
Whether to include default when parsing data NaN value . Depending on whether or not it's coming in na_values, Its behavior is as follows
keep_default_na=True, And it specifies na_values, na_values Will be the same as the default NaN Be parsed together
keep_default_na=True, And no na_values, Only the default NaN
keep_default_na=False, And it specifies na_values, Parse only na_values designated NaN
keep_default_na=False, And no na_values, The string will not be parsed to NaN
Be careful : If na_filter=False, that keep_default_na and na_values Parameters are ignored
na_filter: boolean, The default is True
Detect missing value markers ( Empty string and na_values Value ). In the absence of any NA Data in , Set up na_filter=False It can improve the performance of reading large files
skip_blank_lines: boolean, The default is True
If True, Skip the blank line , It is not interpreted as NaN value
1.5 Date time processingparse_dates: Boolean value 、 List or nested list 、 Dictionaries , The default is False.
If True -> Try to parse the index
If [1, 2, 3] -> Try to 1, 2, 3 Columns resolve to delimited dates
If [[1, 3]] -> take 1, 3 Column resolves to a single date column
If {'foo': [1, 3]} -> take 1, 3 Column as the date and set the column name to foo
infer_datetime_format: Boolean value , The default is False
If set to True And set up parse_dates, Try to infer datetime Format to speed up processing
date_parser: function , The default is None
A function for converting a string sequence into an array of datetime instances . By default dateutil.parser.parser convert ,pandas You will try to call... In three different ways date_parser
Pass one or more arrays (parse_dates Defined columns ) As a parameter ;
take parse_dates String values in defined columns are concatenated into a single array , And pass it on ;
Use one or more strings ( Corresponding to parse_dates Defined columns ) As a parameter , Call... On each line date_parser once .
dayfirst: Boolean value , The default is False
DD/MM Format date
cache_dates: Boolean value , The default is True
If True, Then the unique 、 Converted date cache to apply datetime transformation .
Parsing duplicate date strings , Especially for date strings with time zone offsets , May significantly increase speed .
1.6 iterationiterator: boolean, The default is False
return TextFileReader Object to iterate or use get_chunk() To get the block
1.7 quote 、 Compression and file formatcompression: {'infer', 'gzip', 'bz2', 'zip', 'xz', None, dict}, The default is 'infer'
Used for instant decompression of disk data . If "infer", If filepath_or_buffer Is the file path and starts with ".gz",".bz2",".zip" or ".xz" ending , Then use gzip,bz2,zip or xz decompression , Otherwise, do not decompress .
If you use "zip", be ZIP The file must contain only one data file to read . Set to None It means not decompressing
You can also use a dictionary , The key is method From {'zip', 'gzip', 'bz2'} Choose from . for example
compression={'method': 'gzip', 'compresslevel': 1, 'mtime': 1}thousandsstr, The default is None
The separator of the value in thousands
decimal: str, The default is '.'
decimal point
float_precision: string, The default is None
Appoint C Which converter should the engine use to handle floating point values . The options for a normal converter are None, The options for high-precision converters are high, The bidirectional converter options are round_trip.
quotechar: str ( The length is 1)
A character used to indicate the beginning and end of referenced data . Delimiters in quoted data are ignored
comment: str, The default is None
Used to skip the line beginning with this character , for example , If comment='#', Will skip # Beginning line
encoding: str, The default is None
Set the encoding format
1.8 Error handlingerror_bad_linesboolean, The default is True
By default , Rows with too many fields ( for example , With too many commas csv file ) Exception will be thrown , And will not return any DataFrame.
If set to False, Then these bad lines will be deleted
warn_bad_linesboolean, The default is True
If error_bad_lines=False And warn_bad_lines=True, Each bad line will output a warning
2. Specify the type of data columnYou can indicate the entire DataFrame Or the data type of each column
In [9]: import numpy as npIn [10]: data = "a,b,c,d\n1,2,3,4\n5,6,7,8\n9,10,11"In [11]: print(data)a,b,c,d1,2,3,45,6,7,89,10,11In [12]: df = pd.read_csv(StringIO(data), dtype=object)In [13]: dfOut[13]: a b c d0 1 2 3 41 5 6 7 82 9 10 11 NaNIn [14]: df["a"][0]Out[14]: '1'In [15]: df = pd.read_csv(StringIO(data), dtype={"b": object, "c": np.float64, "d": "Int64"})In [16]: df.dtypesOut[16]: a int64b objectc float64d Int64dtype: objectYou can use read_csv() Of converters Parameters , Unify the data type of a column
In [17]: data = "col_1\n1\n2\n'A'\n4.22"In [18]: df = pd.read_csv(StringIO(data), converters={"col_1": str})In [19]: dfOut[19]: col_10 11 22 'A'3 4.22In [20]: df["col_1"].apply(type).value_counts()Out[20]: <class 'str'> 4Name: col_1, dtype: int64perhaps , You can use... After reading the data to_numeric() Function cast type
In [21]: df2 = pd.read_csv(StringIO(data))In [22]: df2["col_1"] = pd.to_numeric(df2["col_1"], errors="coerce")In [23]: df2Out[23]: col_10 1.001 2.002 NaN3 4.22In [24]: df2["col_1"].apply(type).value_counts()Out[24]: <class 'float'> 4Name: col_1, dtype: int64It converts all valid numeric values to floating point numbers , And resolve the invalid to NaN
Last , How to handle columns with mixed types depends on your specific needs . In the example above , If you just want to convert the abnormal data to NaN, that to_numeric() Maybe the best choice for you .
However , If you want to force all the data , Whatever the type , So use read_csv() Of converters The parameters will be better
Be careful
In some cases , Reading exception data that contains mixed type columns will result in inconsistent data sets .
If you rely on pandas To infer the type of column , The parsing engine will continue to infer the type of the data block , Instead of extrapolating the entire dataset at once .
In [25]: col_1 = list(range(500000)) + ["a", "b"] + list(range(500000))In [26]: df = pd.DataFrame({"col_1": col_1})In [27]: df.to_csv("foo.csv")In [28]: mixed_df = pd.read_csv("foo.csv")In [29]: mixed_df["col_1"].apply(type).value_counts()Out[29]: <class 'int'> 737858<class 'str'> 262144Name: col_1, dtype: int64In [30]: mixed_df["col_1"].dtypeOut[30]: dtype('O')This leads to mixed_df Some blocks of columns contain int type , Other blocks contain str, This is because the data read is of mixed type .
That's all Python pandas Data reading and writing operations IO Tools CSV Details of , More about Python pandas For data reading and writing materials, please pay attention to other relevant articles on the software development network !
 The python project module is packaged and published locally and uploaded to the PyPI official website
The python project module is packaged and published locally and uploaded to the PyPI official website
當完成某個功能模塊開發後
 I used Python to crawl wechat friends. It turns out that they are such people
I used Python to crawl wechat friends. It turns out that they are such people
With the popularity of wechat