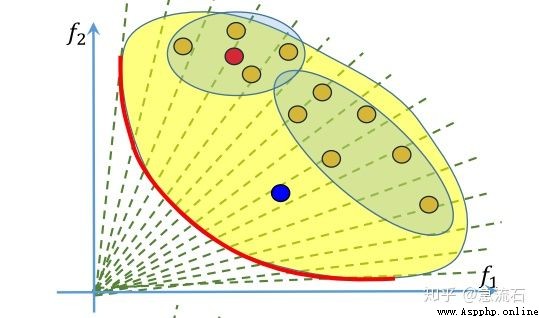
answer : Each solution corresponds to a set of weights , That is, the sub problem , Four dots near the red dot , That is, how can its neighbors be sure ? Determined by weight , At the initialization stage of the algorithm, the neighbors corresponding to each weight are determined , That is, the neighbor subproblem of each subproblem . Weighted neighbors are judged by Euclidean distance . Take the nearest few .
https://www.cnblogs.com/Twobox/p/16408751.html
https://www.zhihu.com/question/263555181?sort=created
Two of the answers were very good
1. Input N m # N Indicates the point density m Represents the problem dimension 1.1 Input T # Means to take the nearest T As neighbors 2. Generate uniformly distributed weight vector 2.1 Calculate the Euler distance between each weight vector 3. The number of weight vectors is : Number of initial population 4. Initial population , Each individual corresponds to the weight one by one 4.1 More weighted distance , Take before T As neighbors person 5. EP = empty # Maintain the best frontier 6. Calculate the initial global optimum Z # Bring each into f1 f2 in , Minimum value z1 z2 7. Start the cycle N generation 7.1 For each individual , Select... In the field 2 Individuals cross mutate , get 2 A new individual 7.1.1 Update global solution z 7.2 Select randomly in the field 2 Individuals , Compare the new with the old # The new individual brings in the sub goal problem , Direct comparison value is enough 7.3 If it's better , Replace the old individual dna 7.4 to update EP # If there is a new solution not received , Combine the new solution with EP Compare each one , Delete the , If the new solution is not dominated by the old solution , Then join in EP
Code implementation design
# analysis Data structures to be maintained : The most recent T Neighbors : Consider using an object list Uniformly distributed weight vector : A two-dimensional ndarray The array can be The weight vector corresponds to the individual : Individual objects , Directly save the weight vector array Distance matrix between weight vectors : Start initialization , constant EP list, The individual inside is the reference of the object z list Set of objective functions ,F list domain list # Interface design class Person attribute: dns: A one-dimensional ndarray weight_vector: A one-dimensional ndarray neighbor: list<Person> o_func:Objective_Function Objective function function: mutation cross_get_two_new_dna: return 2 Duan Xin dna compare# Compare with the offspring accept_new_dna choice_two_person:p1,p2 class Moead_Util attribute: N M T: o_func:Objective_Function pm: Mutation probability EP:[dna1,dna2,..] weight_vectors: Two dimensional array Euler_distance: Two dimensional array pip_size Z:[] # The elements here are one-dimensional ndarray Array , namely dna, Immediate solution function: init_mean_vector: Two dimensional array init_Euler_distance: Two dimensional array init_population:[] init_Z: One dimensional genus pig update_ep update_Z class Objective_Function: attribute: F:[] domain:[[0,1],[],[]] function: get_one_function:Objective_Function
1 import numpy as np 2 3 4 class Person: 5 def __init__(self, dna): 6 self.dna = dna 7 self.weight_vector = None 8 self.neighbor = None 9 self.o_func = None # Objective function 10 11 self.dns_len = len(dna) 12 13 def set_info(self, weight_vector, neighbor, o_func): 14 self.weight_vector = weight_vector 15 self.neighbor = neighbor 16 self.o_func = o_func# Objective function 17 18 def mutation_dna(self, one_dna): 19 i = np.random.randint(0, self.dns_len) 20 low = self.o_func.domain[i][0] 21 high = self.o_func.domain[i][1] 22 new_v = np.random.rand() * (high - low) + low 23 one_dna[i] = new_v 24 return one_dna 25 26 def mutation(self): 27 i = np.random.randint(0, self.dns_len) 28 low = self.o_func.domain[i][0] 29 high = self.o_func.domain[i][1] 30 new_v = np.random.rand() * (high - low) + low 31 self.dna[i] = new_v 32 33 @staticmethod 34 def cross_get_two_new_dna(p1, p2): 35 # A single point of intersection 36 cut_i = np.random.randint(1, p1.dns_len - 1) 37 dna1 = p1.dna.copy() 38 dna2 = p2.dna.copy() 39 temp = dna1[cut_i:].copy() 40 dna1[cut_i:] = dna2[cut_i:] 41 dna2[cut_i:] = temp 42 return dna1, dna2 43 44 def compare(self, son_dna): 45 F = self.o_func.f_funcs 46 f_x_son_dna = [] 47 f_x_self = [] 48 for f in F: 49 f_x_son_dna.append(f(son_dna)) 50 f_x_self.append(f(self.dna)) 51 fit_son_dna = np.array(f_x_son_dna) * self.weight_vector 52 fit_self = np.array(f_x_self) * self.weight_vector 53 return fit_son_dna.sum() - fit_self.sum() 54 55 def accept_new_dna(self, new_dna): 56 self.dna = new_dna 57 58 def choice_two_person(self): 59 neighbor = self.neighbor 60 neighbor_len = len(neighbor) 61 idx = np.random.randint(0, neighbor_len, size=2) 62 p1 = self.neighbor[idx[0]] 63 p2 = self.neighbor[idx[1]] 64 return p1, p2
1 from collections import defaultdict
2
3 import numpy as np
4
5
6 def zdt4_f1(x_list):
7 return x_list[0]
8
9
10 def zdt4_gx(x_list):
11 sum = 1 + 10 * (10 - 1)
12 for i in range(1, 10):
13 sum += x_list[i] ** 2 - 10 * np.cos(4 * np.pi * x_list[i])
14 return sum
15
16
17 def zdt4_f2(x_list):
18 gx_ans = zdt4_gx(x_list)
19 if x_list[0] < 0:
20 print("????: x_list[0] < 0:", x_list[0])
21 if gx_ans < 0:
22 print("gx_ans < 0", gx_ans)
23 if (x_list[0] / gx_ans) <= 0:
24 print("x_list[0] / gx_ans<0:", x_list[0] / gx_ans)
25
26 ans = 1 - np.sqrt(x_list[0] / gx_ans)
27 return ans
28
29 def zdt3_f1(x):
30 return x[0]
31
32
33 def zdt3_gx(x):
34 if x[:].sum() < 0:
35 print(x[1:].sum(), x[1:])
36 ans = 1 + 9 / 29 * x[1:].sum()
37 return ans
38
39
40 def zdt3_f2(x):
41 g = zdt3_gx(x)
42 ans = 1 - np.sqrt(x[0] / g) - (x[0] / g) * np.sin(10 * np.pi * x[0])
43 return ans
44
45
46 class Objective_Function:
47 function_dic = defaultdict(lambda: None)
48
49 def __init__(self, f_funcs, domain):
50 self.f_funcs = f_funcs
51 self.domain = domain
52
53 @staticmethod
54 def get_one_function(name):
55 if Objective_Function.function_dic[name] is not None:
56 return Objective_Function.function_dic[name]
57
58 if name == "zdt4":
59 f_funcs = [zdt4_f1, zdt4_f2]
60 domain = [[0, 1]]
61 for i in range(9):
62 domain.append([-5, 5])
63 Objective_Function.function_dic[name] = Objective_Function(f_funcs, domain)
64 return Objective_Function.function_dic[name]
65
66 if name == "zdt3":
67 f_funcs = [zdt3_f1, zdt3_f2]
68 domain = [[0, 1] for i in range(30)]
69 Objective_Function.function_dic[name] = Objective_Function(f_funcs, domain)
70 return Objective_Function.function_dic[name] 1 import numpy as np
2
3 from GA.MOEAD.Person import Person
4
5
6 def distribution_number(sum, m):
7 # take m Number , The sum of numbers is N
8 if m == 1:
9 return [[sum]]
10 vectors = []
11 for i in range(1, sum - (m - 1) + 1):
12 right_vec = distribution_number(sum - i, m - 1)
13 a = [i]
14 for item in right_vec:
15 vectors.append(a + item)
16 return vectors
17
18
19 class Moead_Util:
20 def __init__(self, N, m, T, o_func, pm):
21 self.N = N
22 self.m = m
23 self.T = T # Neighbor size limit
24 self.o_func = o_func
25 self.pm = pm # Mutation probability
26
27 self.Z = np.zeros(shape=m)
28
29 self.EP = [] # the front
30 self.EP_fx = [] # ep The corresponding target value
31 self.weight_vectors = None # Uniform weight vector
32 self.Euler_distance = None # Euler distance matrix
33 self.pip_size = -1
34
35 self.pop = None
36 # self.pop_dna = None
37
38 def init_mean_vector(self):
39 vectors = distribution_number(self.N + self.m, self.m)
40 vectors = (np.array(vectors) - 1) / self.N
41 self.weight_vectors = vectors
42 self.pip_size = len(vectors)
43 return vectors
44
45 def init_Euler_distance(self):
46 vectors = self.weight_vectors
47 v_len = len(vectors)
48
49 Euler_distance = np.zeros((v_len, v_len))
50 for i in range(v_len):
51 for j in range(v_len):
52 distance = ((vectors[i] - vectors[j]) ** 2).sum()
53 Euler_distance[i][j] = distance
54
55 self.Euler_distance = Euler_distance
56 return Euler_distance
57
58 def init_population(self):
59 pop_size = self.pip_size
60 dna_len = len(self.o_func.domain)
61 pop = []
62 pop_dna = np.random.random(size=(pop_size, dna_len))
63 # Of the original individual dna
64 for i in range(pop_size):
65 pop.append(Person(pop_dna[i]))
66
67 # Of the original individual weight_vector, neighbor, o_func
68 for i in range(pop_size):
69 # weight_vector, neighbor, o_func
70 person = pop[i]
71 distance = self.Euler_distance[i]
72 sort_arg = np.argsort(distance)
73 weight_vector = self.weight_vectors[i]
74 # neighbor = pop[sort_arg][:self.T]
75 neighbor = []
76 for i in range(self.T):
77 neighbor.append(pop[sort_arg[i]])
78
79 o_func = self.o_func
80 person.set_info(weight_vector, neighbor, o_func)
81 self.pop = pop
82 # self.pop_dna = pop_dna
83
84 return pop
85
86 def init_Z(self):
87 Z = np.full(shape=self.m, fill_value=float("inf"))
88 for person in self.pop:
89 for i in range(len(self.o_func.f_funcs)):
90 f = self.o_func.f_funcs[i]
91 # f_x_i: An individual , In the i Value on target
92 f_x_i = f(person.dna)
93 if f_x_i < Z[i]:
94 Z[i] = f_x_i
95
96 self.Z = Z
97
98 def get_fx(self, dna):
99 fx = []
100 for f in self.o_func.f_funcs:
101 fx.append(f(dna))
102 return fx
103
104 def update_ep(self, new_dna):
105 # Combine the new solution with EP Compare each one , Delete the
106 # If the new solution is not dominated by the old solution , The retention
107 new_dna_fx = self.get_fx(new_dna)
108 accept_new = True # Whether to add the new solution to EP
109 # print(f" Ready to start the cycle : EP length {len(self.EP)}")
110 for i in range(len(self.EP) - 1, -1, -1): # Go back and forth
111 old_ep_item = self.EP[i]
112 old_fx = self.EP_fx[i]
113 # old_fx = self.get_fx(old_ep_item)
114 a_b = True # The old ruling line
115 b_a = True # The new rules the old
116 for j in range(len(self.o_func.f_funcs)):
117 if old_fx[j] < new_dna_fx[j]:
118 b_a = False
119 if old_fx[j] > new_dna_fx[j]:
120 a_b = False
121 # T T : fx equal Do not change directly EP
122 # T F : The old rules the new Stay old , Never new , End of cycle .
123 # F T : The new rules the old Stay new , Don't be so old , Continue to cycle
124 # F F : Non dominant relationship Do not operate , Cycle to the next
125 # TF Why end the loop ,FT Why continue the cycle , You can think about
126 if a_b:
127 accept_new = False
128 break
129 if not a_b and b_a:
130 if len(self.EP) <= i:
131 print(len(self.EP), i)
132 del self.EP[i]
133 del self.EP_fx[i]
134 continue
135
136 if accept_new:
137 self.EP.append(new_dna)
138 self.EP_fx.append(new_dna_fx)
139 return self.EP, self.EP_fx
140
141 def update_Z(self, new_dna):
142 new_dna_fx = self.get_fx(new_dna)
143 Z = self.Z
144 for i in range(len(self.o_func.f_funcs)):
145 if new_dna_fx[i] < Z[i]:
146 Z[i] = new_dna_fx[i]
147 return Zimport random
import numpy as np
from GA.MOEAD.Moead_Util import Moead_Util
from GA.MOEAD.Objective_Function import Objective_Function
from GA.MOEAD.Person import Person
import matplotlib.pyplot as plt
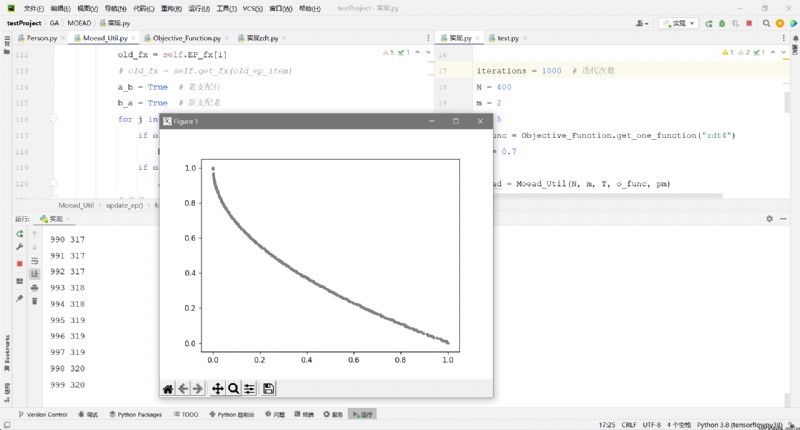
def draw(x, y):
plt.scatter(x, y, s=10, c="grey") # s Size of points c The color of the point alpha transparency
plt.show()
iterations = 1000 # The number of iterations
N = 400
m = 2
T = 40
o_func = Objective_Function.get_one_function("zdt3")
pm = 0.5
moead = Moead_Util(N, m, T, o_func, pm)
moead.init_mean_vector()
moead.init_Euler_distance()
pop = moead.init_population()
moead.init_Z()
for i in range(iterations):
print(i, len(moead.EP))
for person in pop:
p1, p2 = person.choice_two_person()
d1, d2 = Person.cross_get_two_new_dna(p1, p2)
if np.random.rand() < pm:
p1.mutation_dna(d1)
if np.random.rand() < pm:
p1.mutation_dna(d2)
moead.update_Z(d1)
moead.update_Z(d2)
t1, t2 = person.choice_two_person()
if t1.compare(d1) < 0:
t1.accept_new_dna(d1)
moead.update_ep(d1)
if t2.compare(d1) < 0:
t2.accept_new_dna(d2)
moead.update_ep(d1)
# Output result drawing
EP_fx = np.array(moead.EP_fx)
x = EP_fx[:, 0]
y = EP_fx[:, 1]
draw(x, y)