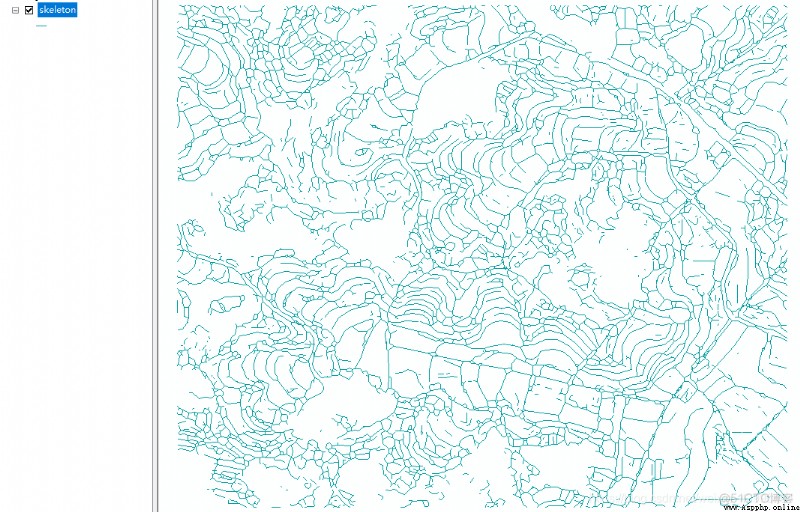
This blog will introduce how to convert grid lines into vector lines . The general process can be divided into , Grid line to graph structure representation , From the point relation of graph structure , Write in line vector .

image2graph.py
# import rdp
# Code Copied From Favyen
import scipy. ndimage
import skimage. morphology
import os
from PIL import Image
import cv2
import math
import numpy
from math import sqrt
import pickle
import gdalTools
def distance( a, b):
return sqrt(( a[ 0] - b[ 0]) * * 2 + ( a[ 1] - b[ 1]) * * 2)
def point_line_distance( point, start, end):
if ( start == end):
return distance( point, start)
else:
n = abs(
( end[ 0] - start[ 0]) * ( start[ 1] - point[ 1]) - ( start[ 0] - point[ 0]) * ( end[ 1] - start[ 1])
)
d = sqrt(
( end[ 0] - start[ 0]) * * 2 + ( end[ 1] - start[ 1]) * * 2
)
return n / d
def rdp( points, epsilon):
"""
Reduces a series of points to a simplified version that loses detail, but
maintains the general shape of the series.
"""
dmax = 0.0
index = 0
for i in range( 1, len( points) - 1):
d = point_line_distance( points[ i], points[ 0], points[ - 1])
if d > dmax:
index = i
dmax = d
if dmax >= epsilon:
results = rdp( points[: index + 1], epsilon)[: - 1] + rdp( points[ index:], epsilon)
else:
results = [ points[ 0], points[ - 1]]
return results
def generateGraph( in_fname):
PADDING = 30
threshold = 1
out_fname = 'graph_gt.pickle'
im_proj, im_geotrans, im_width, im_height, im_data = gdalTools. read_img( in_fname)
im = im_data
im = numpy. array( im)
if len( im. shape) == 3:
print( 'warning: bad shape {}, using first channel only'. format( im. shape))
im = im[:, :, 0]
im = numpy. swapaxes( im, 0, 1)
im = ( im >= threshold)
Image. fromarray( im. astype( 'uint8') * 60). save( "tmp0.png")
im = skimage. morphology. thin( im)
im = im. astype( 'uint8')
Image. fromarray( im * 255). save( "tmp.png")
# extract a graph by placing vertices every THRESHOLD pixels, and at all intersections
vertices = []
edges = set()
def add_edge( src, dst):
if ( src, dst) in edges or ( dst, src) in edges:
return
elif src == dst:
return
edges. add(( src, dst))
point_to_neighbors = {}
q = []
while True:
if len( q) > 0:
lastid, i, j = q. pop()
path = [ vertices[ lastid], ( i, j)]
if im[ i, j] == 0:
continue
point_to_neighbors[( i, j)]. remove( lastid)
if len( point_to_neighbors[( i, j)]) == 0:
del point_to_neighbors[( i, j)]
else:
w = numpy. where( im > 0)
if len( w[ 0]) == 0:
break
i, j = w[ 0][ 0], w[ 1][ 0]
lastid = len( vertices)
vertices. append(( i, j))
path = [( i, j)]
while True:
im[ i, j] = 0
neighbors = []
for oi in [ - 1, 0, 1]:
for oj in [ - 1, 0, 1]:
ni = i + oi
nj = j + oj
if ni >= 0 and ni < im. shape[ 0] and nj >= 0 and nj < im. shape[ 1] and im[ ni, nj] > 0:
neighbors. append(( ni, nj))
if len( neighbors) == 1 and ( i, j) not in point_to_neighbors:
ni, nj = neighbors[ 0]
path. append(( ni, nj))
i, j = ni, nj
else:
if len( path) > 1:
path = rdp( path, 2)
if len( path) > 2:
for point in path[ 1: - 1]:
curid = len( vertices)
vertices. append( point)
add_edge( lastid, curid)
lastid = curid
neighbor_count = len( neighbors) + len( point_to_neighbors. get(( i, j), []))
if neighbor_count == 0 or neighbor_count >= 2:
curid = len( vertices)
vertices. append( path[ - 1])
add_edge( lastid, curid)
lastid = curid
for ni, nj in neighbors:
if ( ni, nj) not in point_to_neighbors:
point_to_neighbors[( ni, nj)] = set()
point_to_neighbors[( ni, nj)]. add( lastid)
q. append(( lastid, ni, nj))
for neighborid in point_to_neighbors. get(( i, j), []):
add_edge( neighborid, lastid)
break
neighbors = {}
# print(vertices)
# with open(out_fname, 'w') as f:
# for vertex in vertices:
# f.write('{} {}\n'.format(vertex[0], vertex[1]))
# f.write('\n')
vertex = vertices
for edge in edges:
nk1 = ( vertex[ edge[ 0]][ 1], vertex[ edge[ 0]][ 0])
nk2 = ( vertex[ edge[ 1]][ 1], vertex[ edge[ 1]][ 0])
if nk1 != nk2:
if nk1 in neighbors:
if nk2 in neighbors[ nk1]:
pass
else:
neighbors[ nk1]. append( nk2)
else:
neighbors[ nk1] = [ nk2]
if nk2 in neighbors:
if nk1 in neighbors[ nk2]:
pass
else:
neighbors[ nk2]. append( nk1)
else:
neighbors[ nk2] = [ nk1]
# f.write('{} {}\n'.format(edge[0], edge[1]))
# f.write('{} {}\n'.format(edge[1], edge[0]))
# print(neighbors)
# pickle.dump(neighbors, open(out_fname, "wb"))
return neighbors
raster2lineShp.py
import gdalTools
import numpy as np
from skimage import morphology
import cv2 as cv
from osgeo import gdalconst, gdal, ogr, osr
import os
from image2graph import *
def imagexy2geo( dataset, col, row):
'''
according to GDAL The six parameter model integrates the coordinates on the image map ( Line number ) Convert to projected coordinates or geographic coordinates ( Coordinate system conversion according to specific data )
:param dataset: GDAL Geographic data
:param row: Line number of pixels
:param col: Column number of pixels
:return: Line number (row, col) Corresponding projection coordinates or geographical coordinates (x, y)
'''
trans = dataset. GetGeoTransform()
px = trans[ 0] + col * trans[ 1] + row * trans[ 2]
py = trans[ 3] + col * trans[ 4] + row * trans[ 5]
return px, py
def raster2LineShp( img_path, strVectorFile):
graph = generateGraph( img_path)
dataset = gdal. Open( img_path)
gdal. SetConfigOption( "GDAL_FILENAME_IS_UTF8", "NO") # In order to support Chinese path
gdal. SetConfigOption( "SHAPE_ENCODING", "CP936") # In order to make the property sheet fields support Chinese
ogr. RegisterAll()
strDriverName = "ESRI Shapefile" # Create data , Here to create ESRI Of shp file
oDriver = ogr. GetDriverByName( strDriverName)
if oDriver == None:
print( "%s The driver is not available !\n", strDriverName)
oDS = oDriver. CreateDataSource( strVectorFile) # create data source
if oDS == None:
print( " create a file 【%s】 Failure !", strVectorFile)
# srs = osr.SpatialReference() # Create a spatial reference
# srs.ImportFromEPSG(4326) # Define a geographic coordinate system WGS1984
srs = osr. SpatialReference(
wkt = dataset. GetProjection()) # I added the output when reading the grid dataset, You don't have to specify projection here , Fully automatic , The above two lines can be annotated , And that proj Parameters can also be removed , Get rid of it yourself
papszLCO = []
# Create Layer , Create a polygon layer ,"TestPolygon"-> Property sheet name
oLayer = oDS. CreateLayer( "TestPolygon", srs, ogr. wkbMultiLineString, papszLCO)
if oLayer == None:
print( " Layer creation failed !\n")
oDefn = oLayer. GetLayerDefn() # Define elements
oFeatureTriangle = ogr. Feature( oDefn)
# Create a single face
for n, v in graph. items():
# if cls > len(cls_dict) - 1:
# continue
for nei in v:
line = ogr. Geometry( ogr. wkbLinearRing) # Build geometry types : Line
nx, ny = n[ 1], n[ 0]
nx, ny = imagexy2geo( dataset, nx, ny)
line. AddPoint( nx, ny) # Add some 01
neix, neiy = nei[ 1], nei[ 0]
neix, neiy = imagexy2geo( dataset, neix, neiy)
line. AddPoint( neix, neiy) # Add some 02
# yard = ogr.Geometry(ogr.wkbLineString) # Build geometry types : polygon
# yard.AddGeometry(line)
# yard.CloseRings()
# geomTriangle = ogr.CreateGeometryFromWkt(str(line)) # Add the closed polygon set to the property sheet
oFeatureTriangle. SetGeometry( line)
oLayer. CreateFeature( oFeatureTriangle)
oDS. Destroy()
if __name__ == '__main__':
rasterPath = './temp/skeleton.tif'
shpPath = './temp/skeleton.shp'
raster2LineShp( rasterPath, shpPath)