Catalog
Preface
One 、 Basic grammar and function
Two 、 Parameter description and code demonstration
1.filepath_or_buffer
2.sep
3.delimiter
4.header
5.names
6.index_col
7.usecols
8.squeeze
9.prefix
10.mangle_dupe_cols
11.dtype
12.engine
13.converters
14.true_values
15.false_values
16.skipinitialspace
17. skiprows
19.nrows
20.na_values
21.keep_default_na
22. na_filter
23.verbose
24.skip_blank_lines
25.parse_dates
26.infer_datetime_format
27.keep_date_col
28.date_parser
29. dayfirst
30.cache_dates
31.iterator
32.chunksize
33.compression
34.thousands
35.decimal
36.lineterminator
37.quotechar
38.quoting
39.doublequote
40.escapechar
41.comment
42.encoding
43.encoding_errors
44.dialect
45. error_bad_lines
46.warn_bad_lines
47.on_bad_lines
48.delim_whitespace
49.low_memory
50.memory_map
51.float_precision
52.storage_options
Focus , Prevent losing , If there is a mistake , Please leave a message , Thank you very much
Pandas It is often used as a data analysis tool library and its own DataFrame Data types do some flexible data conversion 、 Calculation 、 Operation and other complex operations , But they are all built after we get the data from the data source . Therefore, as an interface function for reading data source information, it must have its powerful and convenient ability , When reading different class sources or data of different classes, there are corresponding read Function can be processed one step first , This will reduce a considerable part of our data processing operations . every last read() function , As a data analyst, I personally think everyone should master and be familiar with its corresponding parameters , Corresponding read() Function bloggers have read three articles in detail read_json、read_excel and read_sql:
Pandas obtain SQL database read_sql() Functions and parameters in detail + The sample code
Pandas Handle JSON file read_json() A detailed explanation + Code display
Pandas in read_excel Detailed explanation of the use of function parameters + The sample code
Looking at the entire data source path , The most commonly used data storage objects :SQL、JSON、EXCEL And what to explain in detail this time CSV All over the country . If you can understand the use of the function parameters, you can reduce a lot of subsequent processing DataFrame Data structure code , Only a few... Need to be set read_csv Parameters can be implemented , Therefore, the original intention of this article is to introduce and use this function in detail to achieve the purpose of thorough mastery . I hope readers can ask questions or opinions after reading , The blogger will maintain the blog for a long time and update it in time , Hope you enjoy it .
Here's the desensitized one user_info.csv Document as a presentation :

read_csv The basic syntax format is :
pandas.read_csv(filepath_or_buffer,
sep=NoDefault.no_default,
delimiter=None,
header='infer',
names=NoDefault.no_default,
index_col=None,
usecols=None,
squeeze=None,
prefix=NoDefault.no_default,
mangle_dupe_cols=True,
dtype=None,
engine=None,
converters=None,
true_values=None,
false_values=None,
skipinitialspace=False,
skiprows=None,
skipfooter=0,
nrows=None,
na_values=None,
keep_default_na=True,
na_filter=True,
verbose=False,
skip_blank_lines=True,
parse_dates=None,
infer_datetime_format=False,
keep_date_col=False,
date_parser=None,
dayfirst=False,
cache_dates=True,
iterator=False,
chunksize=None,
compression='infer',
thousands=None,
decimal='.',
lineterminator=None,
quotechar='"',
quoting=0,
doublequote=True,
escapechar=None,
comment=None,
encoding=None,
encoding_errors='strict',
dialect=None,
error_bad_lines=None,
warn_bad_lines=None,
on_bad_lines=None,
delim_whitespace=False,
low_memory=True,
memory_map=False,
float_precision=None,
storage_options=None)You can see that there are quite a few parameters , Compared with read_excel、read_json and read_sql Add up to more . Explains the use of csv Files store data several times as often as other record data files , So about csv There are so many processing parameters for files . It's a good thing , When we will csv Convert file to DataFrame Data reprocessing , You need to write a lot of code to deal with , Providing so many parameters can greatly speed up our efficiency in processing files .
read_csv The basic function of is to csv File to DataFrame Or is it TextParser, It also supports the option of iterating or decomposing files into blocks .
import numpy as np
import pandas as pd
df_csv=pd.read_csv('user_info.csv')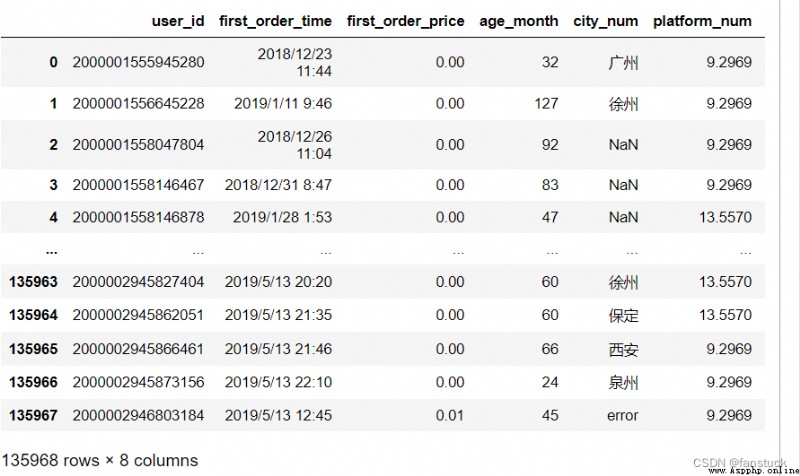
The following is the official document , There are too many words. It is recommended to directly click the directory to see :
pandas.read_csv
First, we will understand the function and function of each parameter one by one , Use the example after understanding the meaning of the parameter .
The type of acceptance :{str, path object or file-like object} character string ( It is usually a file name and can only be read under the directory of the currently running program )、 File path ( Generally, absolute path is recommended )、 Or other data types that can be referenced . Such as url,http This type of .
This parameter specifies the path to the read in file . Any valid string path can be accepted . The string can be URL. Effective URL Include http、ftp、s3、gs And documents . For the file URL, Local files can be :file://localhost/path/to/table.csv.
If you want to pass in a path object ,pandas Accept any path .
For class file objects , We use read() Method reference object , For example, file handles ( For example, through the built-in open function ) or StringIO.
df_csv=pd.read_csv('user_info.csv')df_csv=pd.read_csv(r'C:\Users\10799\test-python\user_info.csv')df_csv=pd.read_csv('C:\\Users\\10799\\test-python\\user_info.csv')The type of acceptance {str, default ‘,’}
csv Generally, the file defaults to ',' To separate each string , and sep Is to specify what type of separator symbol is .
If sep by None, be C The engine cannot automatically detect delimiters , but Python The parsing engine can , It means Python Built in sniffer tool csv The latter will be used and the separator will be automatically detected . Besides , The length exceeds 1 Characters and with “\s+” Different delimiters will be interpreted as regular expressions , And will force the use of Python Parsing engine . Please note that regex Delimiters tend to ignore referenced data .regex Examples of regular expressions :“\r\t”.
df_csv=pd.read_csv(r'C:\Users\10799\test-python\user_info.csv',sep=',')
df_csv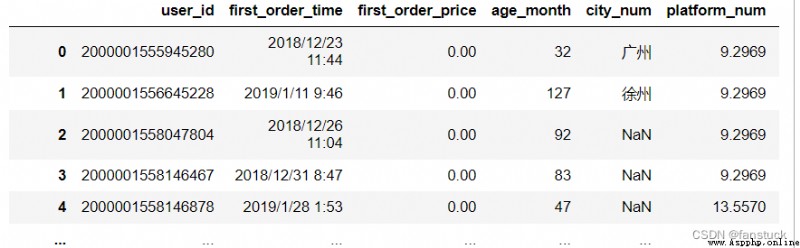
about sep Characters can refer to escape characters :
Escape character describe \( When at the tail ) Line continuation operator \\ Backslash notation \‘ Single quotation marks \” Double quotes \a Ring the bell \b Backspace (Backspace)\e escape \000 empty \n Line break \v Vertical tabs \t Horizontal tabs \r enter \f Change the page \oyy Octal number yy Representative character , for example :\o12 On behalf of the line \xyy Decimal number yy Representative character , for example :\x0a On behalf of the line \other The other characters are output in normal formatdf_csv=pd.read_csv(r'C:\Users\10799\test-python\user_info.csv',sep='/t')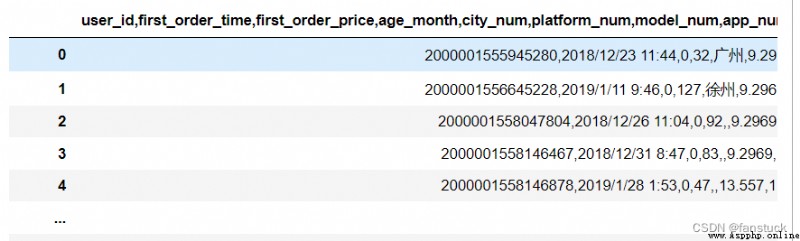
In this way, the experiment will not be repeated , If you are interested, you can try it by yourself .
The type of acceptance :{str, default None}
sep Another name for , Can be set to the desired name , It doesn't make much sense .
df_csv=pd.read_csv(r'C:\Users\10799\test-python\user_info.csv',delimiter=',')
df_csv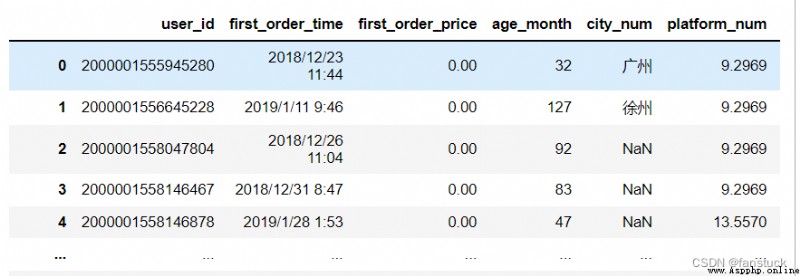
The type of acceptance :{int, list of int, None, default ‘infer’} Integers 、 List of integers 、None. The default is infer
Specify the row number to use as the column name , And the beginning of the data . The default behavior is to infer column names : If no name is passed , Behavior and header=0 identical , And infer column names from the first line of the file , If you explicitly pass the column name , Behavior and header=None identical . Deliver... Explicitly header=0, So that you can replace the existing name . The title can be a list of integers , Specify the row position of multiple indexes on the column , for example [0,1,3]. Unspecified middle lines will be skipped ( for example , Skip... In this example 2). Please note that , If skip_blank_lines=True, This parameter ignores comment lines and empty lines , therefore header=0 Represents the first row of data , Instead of the first line of the file .
df_csv=pd.read_csv(r'C:\Users\10799\test-python\user_info.csv',header=1)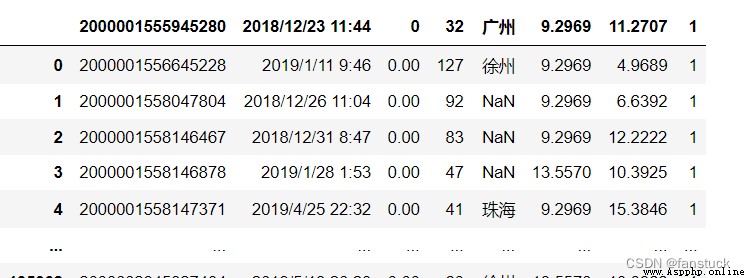
df_csv=pd.read_csv(r'C:\Users\10799\test-python\user_info.csv',header=[0,1,3])In this case . The second case will disappear :
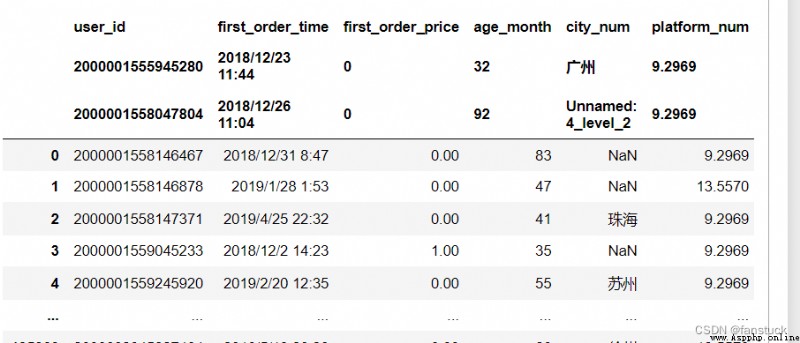
df_csv=pd.read_csv(r'C:\Users\10799\test-python\user_info.csv',header=None)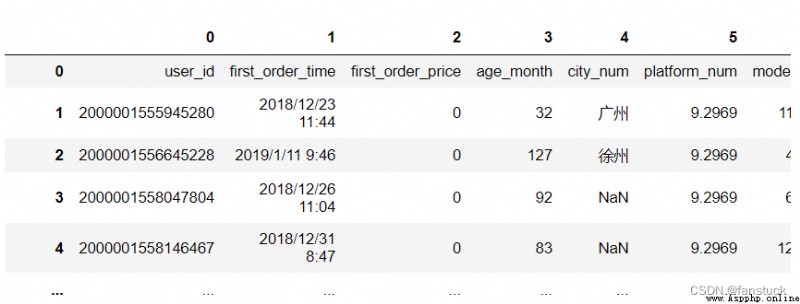
The type of acceptance :{array-like, optional}
Specify the list of column names to use . If the file contains a header line , Should be passed explicitly header=0 To override the column name . Duplicate... Is not allowed in this list .
df_csv=pd.read_csv(r'C:\Users\10799\test-python\user_info.csv',
header=0,names=['id','time','name1','name2','name3','name4','name5','name6'])
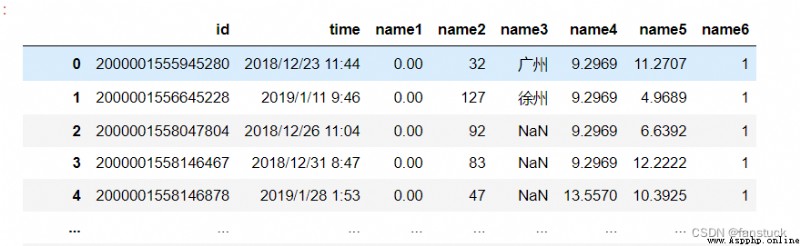
The type of acceptance :{int, str, sequence of int / str, or False, optional} The default is None.
Specify to use as DataFrame Columns of row labels , Given as a string name or column index . If a given int/str Sequence , Use multiple indexes .
Be careful :index_col=False Can be used to force pandas Do not use the first column as an index , for example , When you have a malformed file , When there is a separator at the end of each line .
df_csv=pd.read_csv(r'C:\Users\10799\test-python\user_info.csv',index_col='user_id')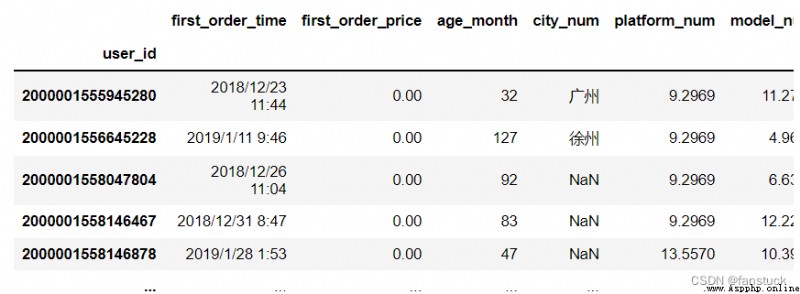
df_csv=pd.read_csv(r'C:\Users\10799\test-python\user_info.csv',index_col=0) 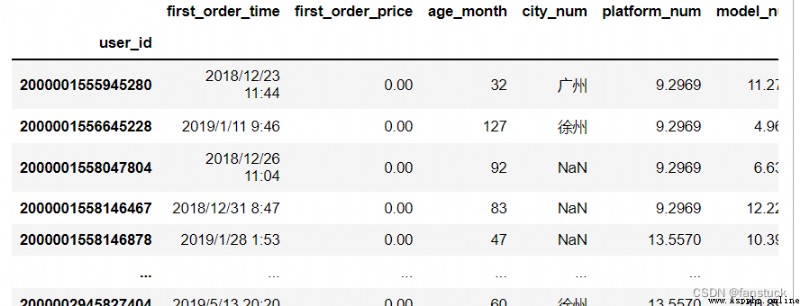
df_csv=pd.read_csv(r'C:\Users\10799\test-python\user_info.csv',index_col=[1,'user_id'])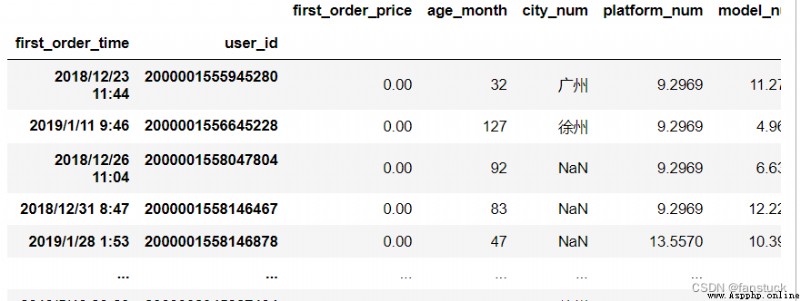
When index_col by False when , Index columns will not be specified .
The type of acceptance :{list-like or callable, optional}
Returns a subset of the column . If it is in list form , Then all elements must be location elements ( That is, the integer index in the document column ) Or a string corresponding to the column name provided by the user in the name or inferred from the document title row . If given names Parameters , The document title line is not considered . for example , image usecols A valid list like parameter should be [0、1、2] or ['foo',bar',baz'].
The specified list order is not taken into account , And the original csv The columns of the file are in the same original order , therefore usecols=[0,1] And [1,0] identical . If callable , The callable function... Will be calculated based on the column name , The return callable function evaluates to True The name of . An example of a valid callable parameter is ['AAA','BBB','DDD' Medium lambda x:x.upper(). Using this parameter can speed up parsing time and reduce memory usage .
df_csv=pd.read_csv(r'C:\Users\10799\test-python\user_info.csv',usecols=['age_month'])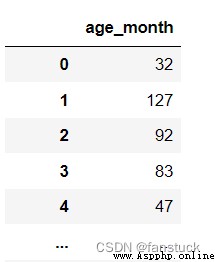
df_csv=pd.read_csv(r'C:\Users\10799\test-python\user_info.csv',usecols=[0,2])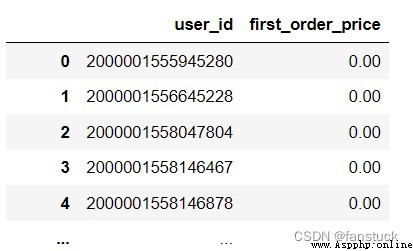
df_csv=pd.read_csv(r'C:\Users\10799\test-python\user_info.csv',
header=0,
names=['id','time','name1','name2','name3','name4','name5','name6'],
usecols=['id','name1']) 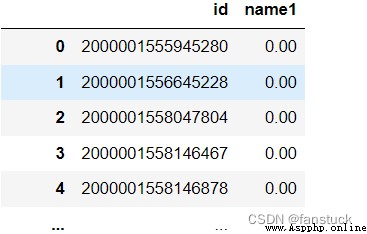
df_csv=pd.read_csv(r'C:\Users\10799\test-python\user_info.csv',
usecols=['age_month','first_order_time']) 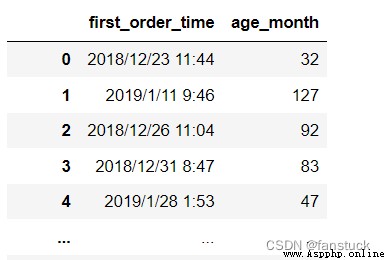
The given list is visible but the resulting DataFrame The order of is not changed according to the order of the given list .
The type of acceptance :{bool, default False}
If the parsed data contains only one column , It returns a sequence .
df_csv=pd.read_csv('user_info.csv',usecols=['age_month'],squeeze=True) 
The type of acceptance :{str, optional}
Prefix added to the column number when there is no title , for example “X” Express X0,X1.
df_csv=pd.read_csv(r'C:\Users\10799\test-python\user_info.csv',header=None,prefix='name')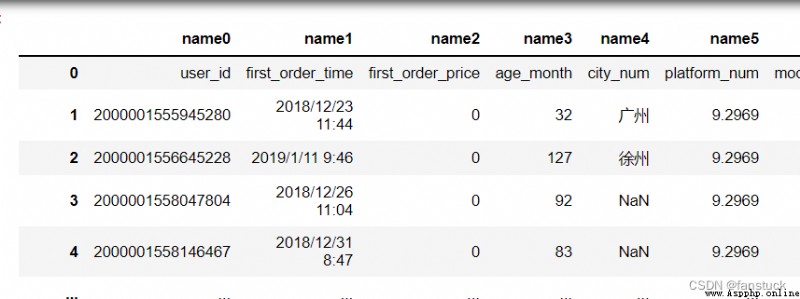
The type of acceptance :{bool, default True}
The repeating column will be specified as “X”、“X.1”、“X.N”, instead of “X”…“X”. If there is a duplicate name in the column , Pass in False Will cause the data to be covered .
df_csv=pd.read_csv(r'C:\Users\10799\test-python\user_info.csv',header=0,names=['id','time','name1','name2','name3','name4','name5','name6'],mangle_dupe_cols=False)ValueError: Setting mangle_dupe_cols=False is not supported yet
Set to False Time is not yet supported .
The type of acceptance :{Type name or dict of column -> type, optional}
Specify the data type of the data or column . for example :{‘a’: np.float64, ‘b’: np.int32, ‘c’: ‘Int64’} .
take str or object With the right na_values Settings are used together , To preserve rather than interpret data types . If a converter is specified , Will be applied INSTEAD Instead of data type conversion .
df_csv=pd.read_csv('user_info.csv')
df_csv.dtypes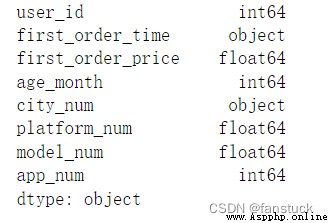
df_csv=pd.read_csv(r'C:\Users\10799\test-python\user_info.csv',dtype={'user_id':'str'})
df_csv.dtypes 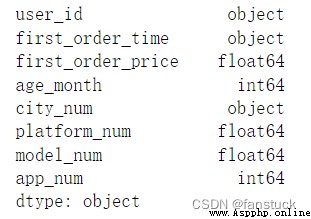
The type of acceptance :{‘c’, ‘python’, ‘pyarrow’}
Specify the parser engine to use .C and pyarrow The engine is faster , and python The current functions of the engine are more complete . Multithreading currently consists only of pyarrow Engine support .
time_start=time.time()
df_csv=pd.read_csv(r'C:\Users\10799\test-python\user_info.csv',engine='python')
time_end=time.time()
print('time cost',time_end-time_start,'s')time cost 1.0299623012542725 s
time_start=time.time()
df_csv=pd.read_csv(r'C:\Users\10799\test-python\user_info.csv',engine='c')
time_end=time.time()
print('time cost',time_end-time_start,'s')time cost 0.20297026634216309 s
The type of acceptance :{dict, optional}
Function for converting values in some columns . Keys can be integers or column labels .
df_csv=pd.read_csv(r'C:\Users\10799\test-python\user_info.csv',converters={'city_num':lambda x:x})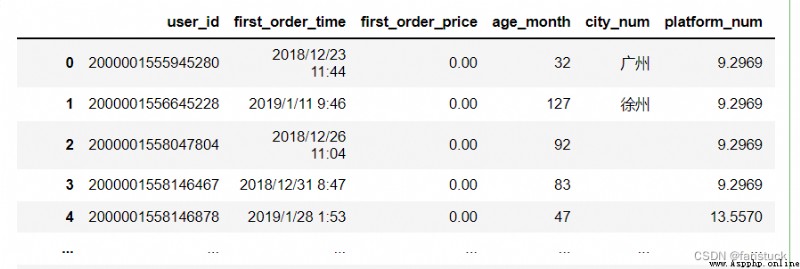
The type of acceptance :{list, optional}
Specifies that the list should be treated as true , When read csv When there are too many, this parameter will have BUG, I can't read it :
df_csv=pd.read_csv('user_info.csv',true_values=[' Guangzhou ']) Guangzhou has not changed , I also tried a lot of values, but it didn't work , The logic of estimating internal parameters is not suitable for large quantities of csv data .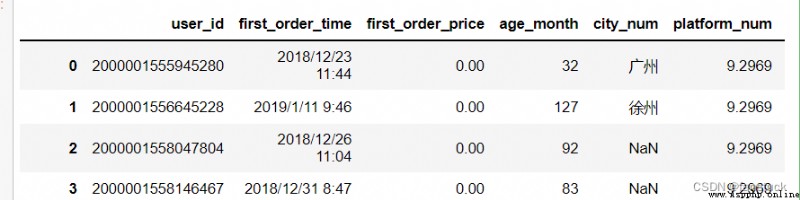
Just run a simple one :
from io import StringIO
data = ('a,b,c\n1,Yes,2\n3,No,4')
pd.read_csv(StringIO(data),
true_values=['Yes'], false_values=['No']) 
It is not recommended that you use this parameter to adjust TRUE, Write your own lambda Function or replace It's the same , This will cause problems in the overall situation . Only when all the data in a column appears in true_values + false_values Inside , Will be replaced .
The type of acceptance :{list, optional}
Specify that the list be treated as false . Same as the previous parameter , It belongs to useless parameter , Pit man 、 skip . Only when all the data in a column appears in true_values + false_values Inside , Will be replaced .
The type of acceptance :{bool, default False}
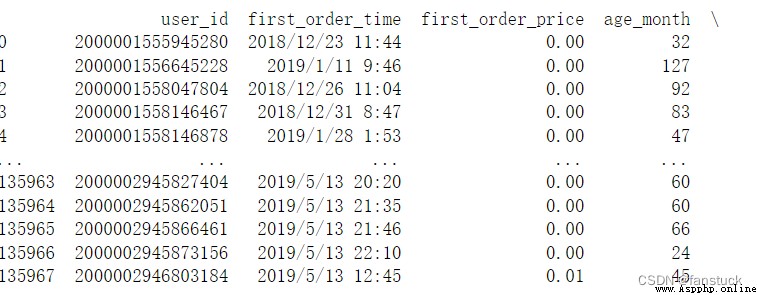
Skip spaces after delimiters .
df_csv=pd.read_csv('user_info.csv',skipinitialspace=True)Do not have what difference :
The type of acceptance :{list-like,int or callable, Optional }
Specify the line number to skip at the beginning of the file (0 Initial index ) Or the number of lines to skip (int).
If callable , Callable functions will be calculated based on the row index , If this line should be skipped , Then return to True, Otherwise return to False. An example of a valid callable parameter is [0,2] Medium lambda x:x.
df_csv=pd.read_csv('user_info.csv',skiprows=[0,1,2,3])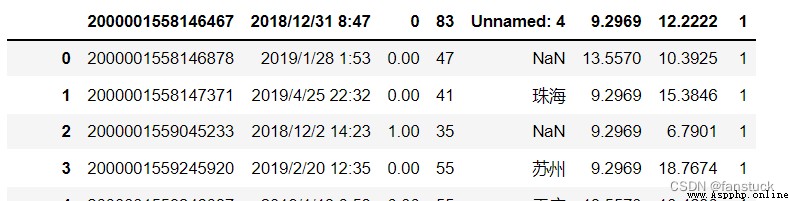
For example, select rows with even rows :
df_csv=pd.read_csv('user_info.csv',skiprows=lambda x :x%2==0)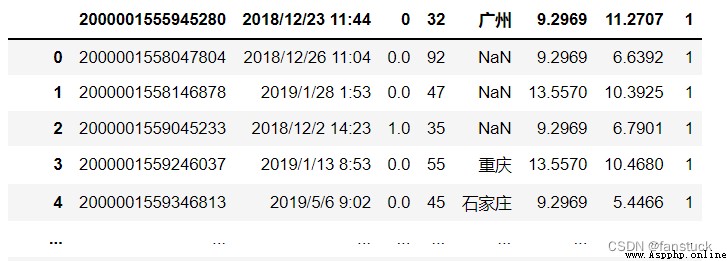 18.skipfooter
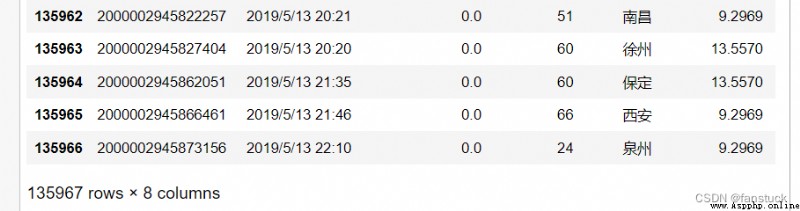
18.skipfooterThe type of acceptance :{int, default 0}
Appoint Number of lines at the bottom of the file to skip (engine='c' I won't support it ).
df_csv=pd.read_csv('user_info.csv',skipfooter=1)Skip the specified number of rows at the bottom :
The type of acceptance :{int, optional}
Specify the number of file lines to read . For reading large files .
df_csv=pd.read_csv('user_info.csv',nrows=50)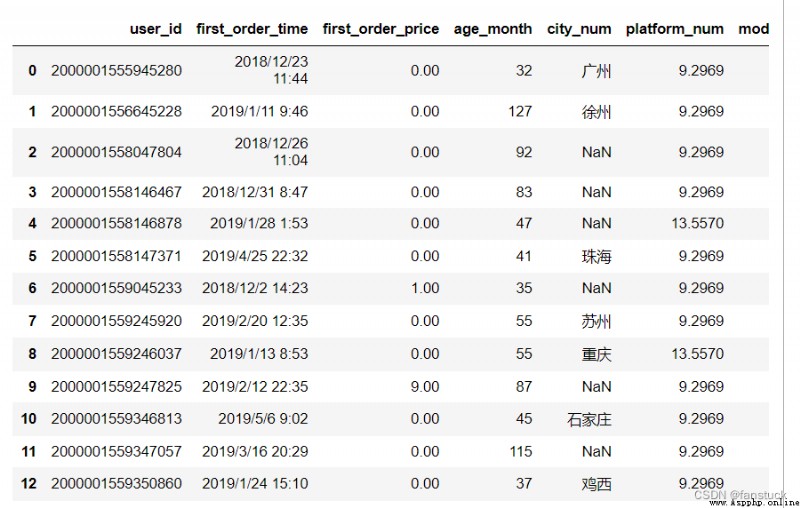
The type of acceptance :{scalar, str, list-like, or dict, optional}
To identify as NA/NaN Other strings . If dict adopt , Specify each column NA value . By default , The following values are interpreted as NaN: ‘’, ‘#N/A’, ‘#N/A N/A’, ‘#NA’, ‘-1.#IND’, ‘-1.#QNAN’, ‘-NaN’, ‘-nan’, ‘1.#IND’, ‘1.#QNAN’, ‘<NA>’, ‘N/A’, ‘NA’, ‘NULL’, ‘NaN’, ‘n/a’, ‘nan’, ‘null’.
df_csv=pd.read_csv('user_info.csv',na_values='0')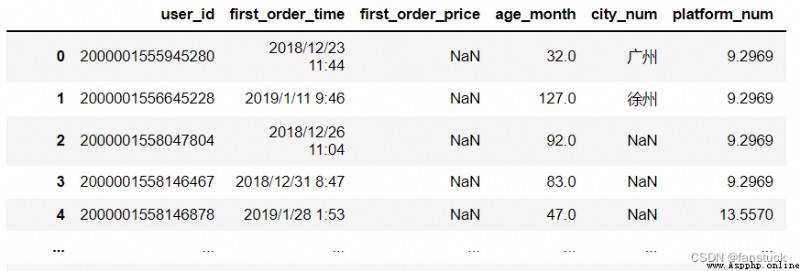
df_csv=pd.read_csv('user_info.csv',na_values=['0','32'])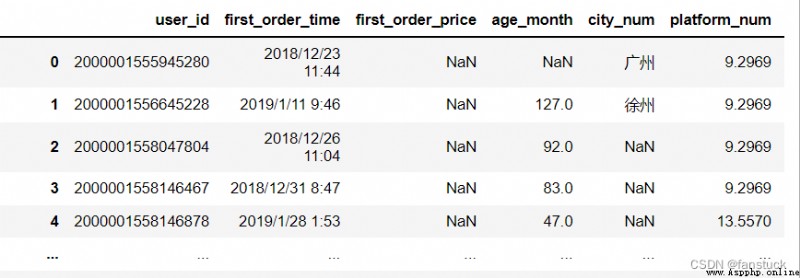
The type of acceptance :{bool, default True}
Whether to include the default when parsing data NaN value . Depending on whether or not it's coming in na_values, The behavior is as follows :
Be careful , If na_filter As False Pass in ,keep_default_na and na_values Parameters are ignored .
df_csv=pd.read_csv('user_info.csv',keep_default_na=False)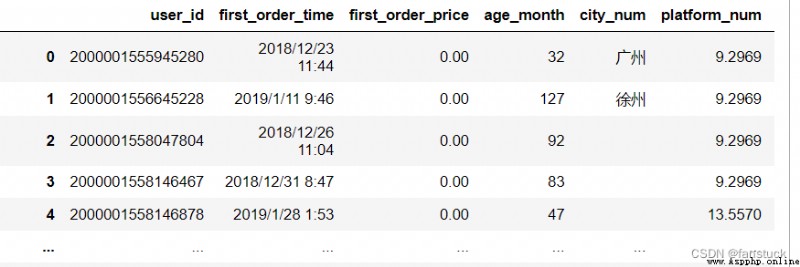
The type of acceptance :{bool, default True}
Detect missing value tags ( Empty string and na_values Value ). In the absence of any NAs Data in , Pass on na_filter=False It can improve the performance of reading large files .
This parameter is used for tuning , Many stored data have null values , So the default is True appropriate .
The type of acceptance :{bool, default False}
Print the output information of various parsers , Can indicate that... Is placed in a non numeric column NA The amount of value .
df_csv=pd.read_csv('user_info.csv',verbose=True) 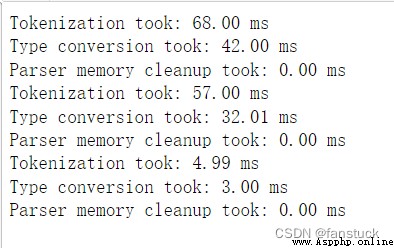
The type of acceptance :{bool, default True}
If True, Skip the blank line , It is not interpreted as NaN value . Otherwise it would be NaN
df_csv=pd.read_csv('user_info.csv',skip_blank_lines=False)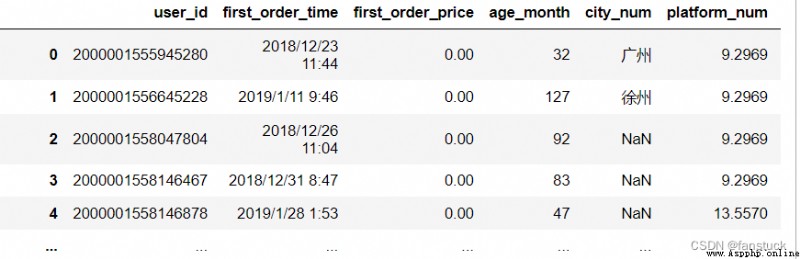
The type of acceptance :{bool or list of int or names or list of lists or dict, default False}
Parameter selection functions are as follows :
If a column or index cannot be represented as datetimes Array , For example, due to non analyzable values or mixing of time zones , Then the column or index will be returned intact as the object data type . For nonstandard datetime analysis ,pd.read_csv Subsequent processing uses to_datetime. To resolve the index or column of the mixed time zone , Please put date_parser Specified as partially applied pandas.to_datetime() Use utc=True. For more information , See using mixed time zone resolution CSV:https://pandas.pydata.org/pandas-docs/stable/user_guide/io.html#io-csv-mixed-timezones.
Be careful : There is iso8601 Quick path to format date .
df_csv=pd.read_csv('user_info.csv',parse_dates=['first_order_time']) 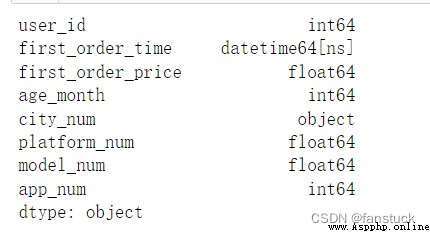
df_csv.dtypes 
df_csv=pd.read_csv('user_info.csv',parse_dates=[1])
df_csv.dtypes 
The type of acceptance :{bool, default False}
If enabled True and parse_dates,pandas Will try to infer... In the column datetime The format of the string , If we can infer , Switch to a faster parsing method . In some cases , This can speed up parsing 5-10 times .
time_start=time.time()
df_csv=pd.read_csv('user_info.csv',parse_dates=[1])
time_end=time.time()
print('time cost',time_end-time_start,'s')
df_csv.dtypes 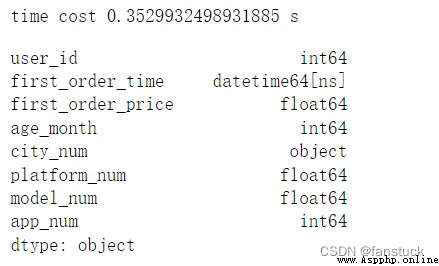
time_start=time.time()
df_csv=pd.read_csv('user_info.csv',parse_dates=[1],infer_datetime_format='%Y/%m/%d %H:%M')
time_end=time.time()
print('time cost',time_end-time_start,'s')
df_csv 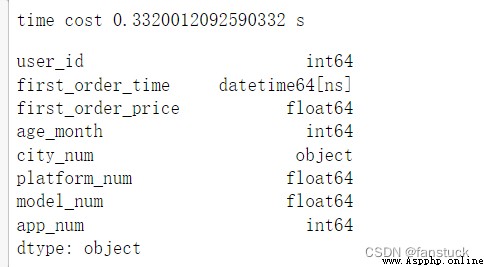
The type of acceptance :{bool, default False}
If True and parse_dates Specify to combine multiple columns , Then keep the original column .
Because if multiple columns specify different time units , After merging, there will be no original columns , Appoint keep_date_col by True when , They will remain .
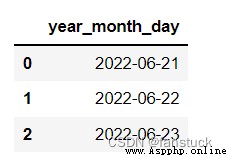
from io import StringIO
data = ('year,month,day\n2022,6,21\n2022,6,22\n2022,6,23')
pd.read_csv(StringIO(data),parse_dates=[[0,1,2]]
)
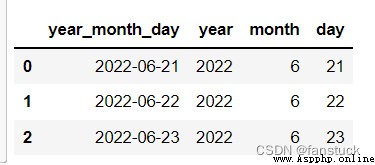
from io import StringIO
data = ('year,month,day\n2022,6,21\n2022,6,22\n2022,6,23')
pd.read_csv(StringIO(data),parse_dates=[[0,1,2]],keep_date_col=True
)
The type of acceptance :{function, optional}
Specify the function , Used to convert a character string column sequence to datetime Instance array . By default dateutil.parser.parser To switch .Pandas You will try to call... In three different ways date_parser, If something unusual happens , Will move on to the next way :1) Pass one or more arrays ( from parse_dates Definition ) As a parameter ;2) take parse_dates String values in defined columns are concatenated ( Press the line ) Into an array , And pass the array ; and 3) Use one or more strings ( Corresponding to parse_dates Defined columns ) As a parameter , Call once for each row parse_dates.
Generally speaking, it will be used in read_csv Convert to DataFrame after , Handle datetime Then write the function , But with this parameter, you can directly process the value of the parameter with time after writing the user-defined function in the early stage .
from io import StringIO
from datetime import datetime
def dele_date(dateframe):
for x in dateframe:
x=pd.to_datetime(x,format='%Y/%m/%d %H:%M')
x.strftime('%m/%d/%Y')
return x
df_csv=pd.read_csv('user_info.csv',parse_dates=['first_order_time'],date_parser=dele_date)
df_csvThe type of acceptance :{bool, default False}
Japan / Month format date , International and European formats .
df_csv=pd.read_csv('user_info.csv',parse_dates=['first_order_time'],dayfirst=True)
df_csv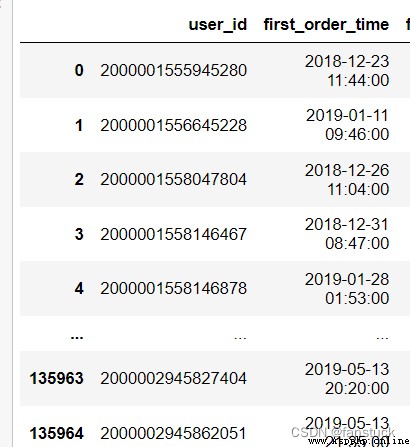
It doesn't work , What form should it be or what form , Unless it is DD/MM Format is useful , Not very useful .
The type of acceptance :{bool, default True}
If True, Use a unique converted date cache to apply datetime conversion . When parsing duplicate date strings , Especially for date strings with time zone offsets , There may be a significant acceleration .
Optimization parameters , To speed up .
The type of acceptance :{bool, default False}
return TextFileReader Object to iterate or get blocks get_chunk().
df_csv=pd.read_csv('user_info.csv',iterator=True)
print(df_csv)<pandas.io.parsers.readers.TextFileReader object at 0x000002624BBA5848>
The type of acceptance :{int, optional}
Returns the for iteration TextFileReader object . of iterator and chunksize For more information , see also IO Tools documentation .
Functional functions , Specify to convert to TextFileReader The number of pieces .
The type of acceptance :{str or dict, default ‘infer’}
Used to decompress disk data in real time . If “infer” and “%s” Similar to path , Detect compression from the following extensions :'.gz','.bz2’,”.zip“,”.xz’, or ’.zst’( Otherwise there is no compression ). If you use “zip”,zip The file must contain only one data file to be read in . Set to “None” Indicates no decompression . It can also be a dict, Middle key “method” Set to {'zip',gzip',bz2',zstd} One of , Other key value pairs are forwarded to zipfile.ZipFile,gzip.gzip file ,bz2.BZ2 File or zstandard.ZstdDecompressor. for example , You can use a custom compression dictionary to Zstandard Decompress and pass the following :compression={'method':'zstd','dict_data':my_compression_dict}.
df_csv=pd.read_csv('user_info.csv',compression=None)
df_csv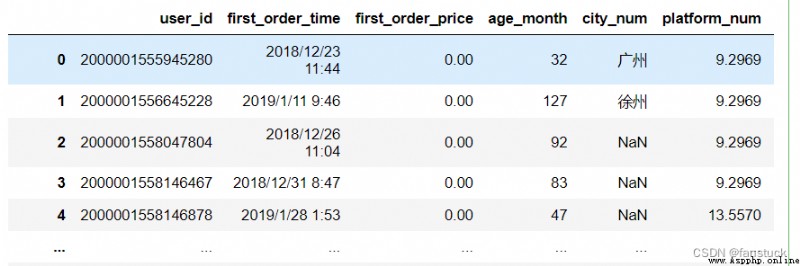
The type of acceptance :{str, optional}
Thousand separator .
The type of acceptance :{str, default ‘.’}
Characters to be recognized as decimal points ( for example , For European data use “,”.)
It's usually float All data are decimal points , I think this parameter may be used for encryption .
The type of acceptance :{str (length 1), optional}
Characters split the file into lines . Only on C The parser is valid . Set to engine by C:
df_csv=pd.read_csv('user_info.csv',engine='c',lineterminator='2')
df_csvThe type of acceptance :{str (length 1), optional}
A character used to indicate the beginning and end of a reference item . Quoted items can contain delimiters , It will be ignored .
The type of acceptance :{int or csv.QUOTE_* instance, default 0}
df_csv=pd.read_csv('user_info.csv',quotechar = '"')
df_csv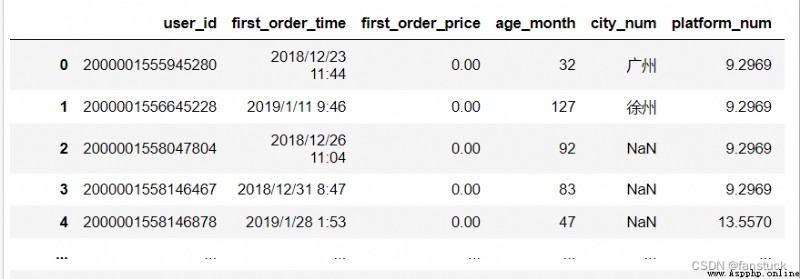
Every csv Control field reference behavior .QUOTE_* Constant . Use QUOTE\u MINIMAL(0)、QUOTE\u ALL(1)、QUOTE_NONNERIAL(2) or QUOTE_NONE(3) One of .
I feel that the following parameters are added temporarily , It's hard to meet ordinary business needs .
The type of acceptance :{bool, default True}
If you specify quotechar And Quoteching No QUOTE_NONE, Please indicate whether two consecutive in the field quotechar The element is interpreted as a single quotechar Elements .
df_csv=pd.read_csv('user_info.csv',quotechar='"', doublequote=True)
df_csv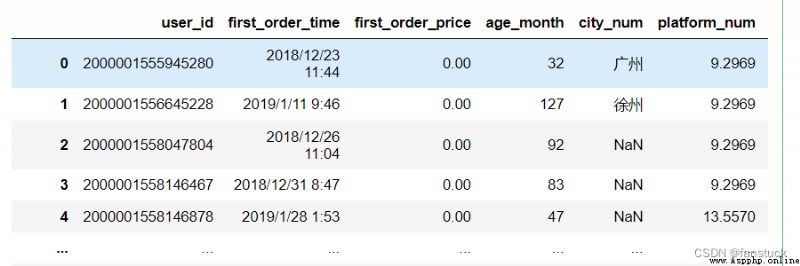
The type of acceptance :{str (length 1), optional}
A string used to escape other characters . When quoting by QUOTE_NONE when , Specifies an undelimited value for a character to cause .
The type of acceptance :{str, optional}
Indicates that the rest of the line should not be parsed . If you find... At the beginning of a line , This line is completely ignored . This parameter must be a single character . Same as blank line ( as long as skip_blank_lines=True), Fully annotated lines are ignored by the parameter header , Rather than being skiprows Ignore . for example , If the note =“#”, The analysis is titled 0 Of #empty\na、b、c\n1、2、3 Will lead to “a、b、c” Be treated as a title .
df_csv=pd.read_csv('user_info.csv',sep=',', comment='#', skiprows=1)
df_csv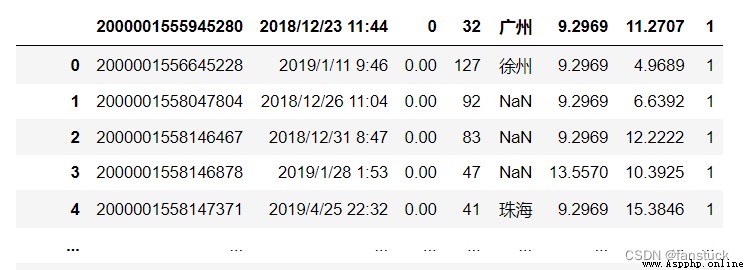
The type of acceptance :{str, optional}
read / Write for UTF The coding ( for example “UTF-8”).
The type of acceptance :{str, optional, default “strict”}
Handling encoding errors .
The type of acceptance :{str or csv.Dialect, optional}
Provided , This parameter will override the values of the following parameters ( Default or non default ):delimiter、doublequote、escapechar、skipinitialspace、quotechar and quoting. If you need to override the value , Will send out ParserWarning. Please see the csv. If no specific language is specified , If sep Larger than one character is ignored . Dialect documentation for more details .
The type of acceptance :{bool, optional, default None}
By default , Rows with too many fields ( for example , Too many commas csv That's ok ) Exception will be thrown , And will not return DataFrame. If False, These... Will be deleted from the returned data frame “ Wrong line ”. This is a good way to handle erroneous data :
The type of acceptance :{bool, optional, default None}
If error_bad_lines by False,warn_bad_lines by True, Each... Will be output “bad line” Warning of .
The type of acceptance :{{‘error’, ‘warn’, ‘skip’} or callable, default ‘error’}
Specify the error line encountered ( Rows with too many fields ) Is the operation to be performed . The allowed values are :
df_csv=pd.read_csv('http://localhost:8889/edit/test-python/user_info.csv',sep=',',on_bad_lines='skip')
df_csv
The type of acceptance :{bool, default False}
Specifies whether spaces will be ( for example “.” or “”) Used as a sep. It's equivalent to setting sep=“\s+”. If this option is set to True, Should not be delimiter Parameter passes in anything .
df_csv=pd.read_csv('user_info.csv',delim_whitespace=True)
df_csv
Here is a space in the time that causes all to separate . Set up sep=“\s+” It's the same .
The type of acceptance :{bool, default True}
Process files internally in blocks , This reduces memory usage during parsing , But it may be a mixed type inference . Make sure there are no mixed types , Please set up False, Or use dtype Parameter specified type . Please note that , The entire file is read into a single data frame , No matter how , Please use chunksize or iterator Parameters return data in blocks .( Only on C The parser is valid ).
The type of acceptance :{bool, default False}
If filepath_or_buffer Provides filepath, Please map the file object directly to memory , And access data directly from memory . Use this option to improve performance , Because there is no longer any I/O expenses .
The type of acceptance :{str, optional}
Appoint C Which converter should the engine use for floating point values . The options for a normal converter are none or “ high ”, The options for the original low precision panda converter are “ Tradition ”, The options for the round trip converter are “ Back and forth ”.
Additional options that make sense for specific storage connections , For example, host 、 port 、 user name 、 Password etc. . about HTTP(S)URL, Key value pairs are forwarded to as header options urllib. For others URL( for example , With “s3://” and “gcs://”) start ), Forward key value pairs to fsspec. For more details , see also fsspec and urllib.
That's what this issue is all about . I am a fanstuck , If you have any questions, please leave a message for discussion , See you next time .