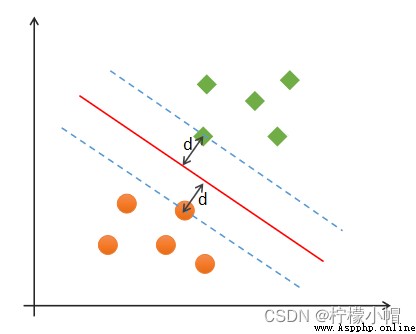
(1) correctness : Most samples can be classified correctly ;
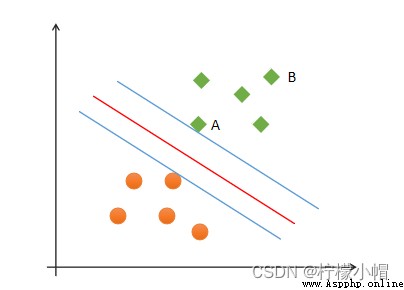
(2) Security : Support vector , That is, the distance between the samples closest to the classification boundary is the farthest ;
(3) Fairness : The distance between the support vector and the classification boundary is equal ;
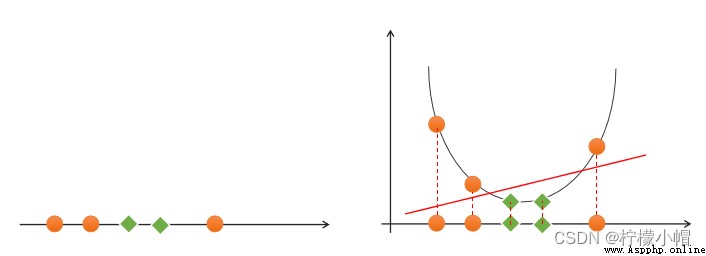
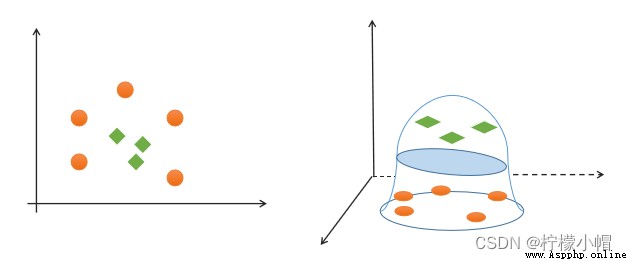
(4) simplicity : Using linear equations ( A straight line 、 Plane ) Represents the classification boundary , Also called split hyperplane . If linear division cannot be made in the original dimension , Then through the dimension lifting transformation , Seek linear partition hyperplane in higher dimensional space . The transformation from low latitude space to high latitude space is carried out by kernel function .
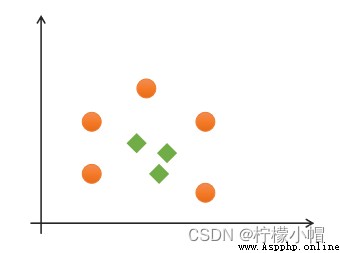
If a set of samples can use a linear function to correctly classify the samples , Call these data samples linearly separable . So what is a linear function ? It's a straight line in two-dimensional space , It's a plane in three-dimensional space , And so on , If you don't consider the dimension of space , Such linear functions are collectively referred to as hyperplanes .
If a set of samples , Unable to find a linear function to correctly classify the samples , These samples are said to be linearly nonseparable . The following is an example of one-dimensional linear indivisibility :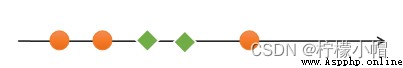
# Support vector machine example
import numpy as np
import sklearn.model_selection as ms
import sklearn.svm as svm
import sklearn.metrics as sm
import matplotlib.pyplot as mp
x, y = [], []
with open("../data/multiple2.txt", "r") as f:
for line in f.readlines():
data = [float(substr) for substr in line.split(",")]
x.append(data[:-1]) # Input
y.append(data[-1]) # Output
# List to array
x = np.array(x)
y = np.array(y, dtype=int)
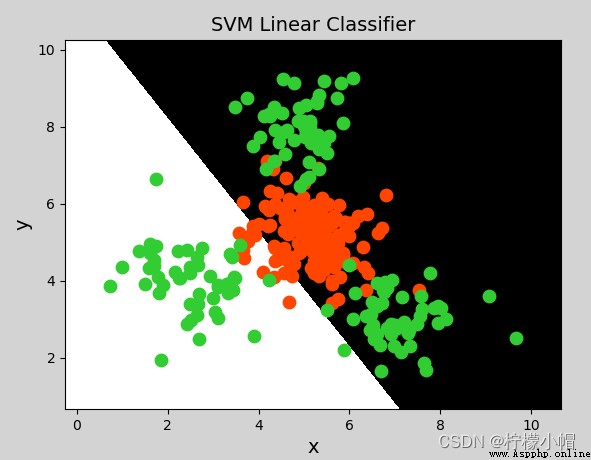
# Linear kernel support vector machine classifier
model = svm.SVC(kernel="linear") # Linear kernel function
# model = svm.SVC(kernel="poly", degree=3) # Polynomial kernel function
# print("gamma:", model.gamma)
# Radial basis function kernel support vector machine classifier
# model = svm.SVC(kernel="rbf",
# gamma=0.01, # Standard deviation of probability density
# C=200) # Probability intensity
model.fit(x, y)
# Calculate drawing boundaries
l, r, h = x[:, 0].min() - 1, x[:, 0].max() + 1, 0.005
b, t, v = x[:, 1].min() - 1, x[:, 1].max() + 1, 0.005
# Generate grid matrix
grid_x = np.meshgrid(np.arange(l, r, h), np.arange(b, t, v))
flat_x = np.c_[grid_x[0].ravel(), grid_x[1].ravel()] # Merge
flat_y = model.predict(flat_x) # According to the grid matrix prediction classification
grid_y = flat_y.reshape(grid_x[0].shape) # Restore shape
mp.figure("SVM Classifier", facecolor="lightgray")
mp.title("SVM Classifier", fontsize=14)
mp.xlabel("x", fontsize=14)
mp.ylabel("y", fontsize=14)
mp.tick_params(labelsize=10)
mp.pcolormesh(grid_x[0], grid_x[1], grid_y, cmap="gray")
C0, C1 = (y == 0), (y == 1)
mp.scatter(x[C0][:, 0], x[C0][:, 1], c="orangered", s=80)
mp.scatter(x[C1][:, 0], x[C1][:, 1], c="limegreen", s=80)
mp.show()
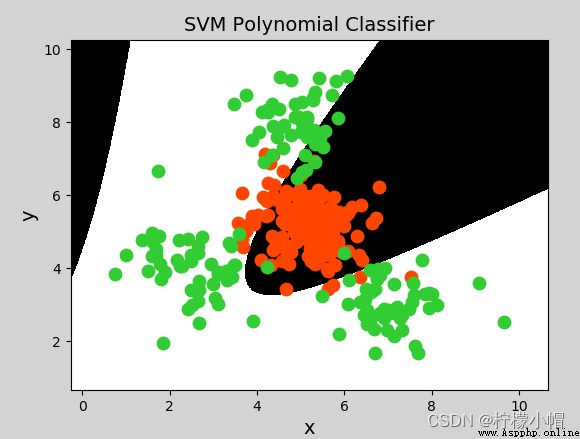
y = x 1 + x 2 y = x 1 2 + 2 x 1 x 2 + x 2 2 y = x 1 3 + 3 x 1 2 x 2 + 3 x 1 x 2 2 + x 2 3 y = x_1 + x_2\\ y = x_1^2 + 2x_1x_2+x_2^2\\ y=x_1^3 + 3x_1^2x_2 + 3x_1x_2^2 + x_2^3 y=x1+x2y=x12+2x1x2+x22y=x13+3x12x2+3x1x22+x23
model = svm.SVC(kernel="poly", degree=3) # Polynomial kernel function
# Radial basis function kernel support vector machine classifier
model = svm.SVC(kernel="rbf",
gamma=0.01, # Standard deviation of probability density
C=600) # Probability intensity , The greater the value, the less tolerance for misclassification , The higher the classification accuracy , But the worse the generalization ability ; The smaller the value , The greater the tolerance for misclassification , But the generalization ability is strong
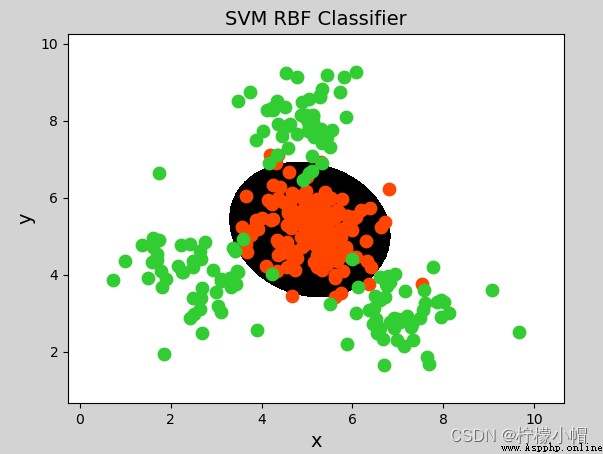
model = svm.SVC(kernel='linear')
model.fit(train_x, train_y)
# Support vector machine classifier based on linear kernel function
model = svm.SVC(kernel='poly', degree=3)
model.fit(train_x, train_y)
# Support vector machine classifier based on radial basis kernel function
# C: Regular intensity
# gamma:'rbf','poly' and 'sigmoid' The kernel function of . The higher the gamma value , The training data set will be accurately fitted , It may lead to over fitting problems .
model = svm.SVC(kernel='rbf', C=600, gamma=0.01)
model.fit(train_x, train_y)
(1) Support vector machine is a binary classification model
(2) Support vector machine finds the optimal linear model as the classification boundary
(3) Boundary requirements : correctness 、 Fairness 、 Security 、 simplicity
(4) Linear nonseparable problem can be transformed into linear separable problem by kernel function , Kernel functions include : Linear kernel function 、 Polynomial kernel function 、 Radial basis function
(5) Support vector machine is suitable for the classification of a small number of samples
import sklearn.model_selection as ms
params =
[{
'kernel':['linear'], 'C':[1, 10, 100, 1000]},
{
'kernel':['poly'], 'C':[1], 'degree':[2, 3]},
{
'kernel':['rbf'], 'C':[1,10,100], 'gamma':[1, 0.1, 0.01]}]
model = ms.GridSearchCV( Model , params, cv= Number of cross validation )
model.fit( Input set , Output set )
# Get each parameter combination of grid search
model.cv_results_['params']
# Get the average test score corresponding to each parameter combination of grid search
model.cv_results_['mean_test_score']
# Get the best parameters
model.best_params_
model.best_score_
model.best_estimator_
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
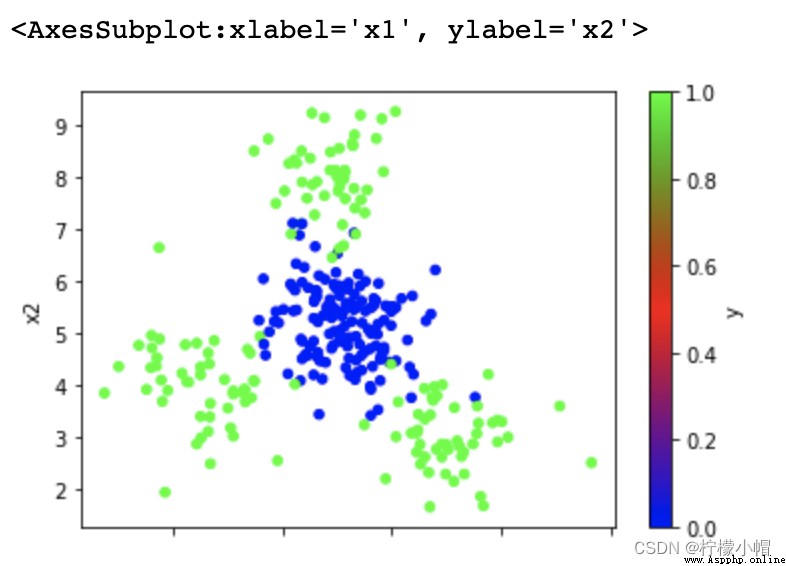
data = pd.read_csv('multiple2.txt', header=None, names=['x1', 'x2', 'y'])
data.plot.scatter(x='x1', y='x2', c='y', cmap='brg')
import sklearn.model_selection as ms
import sklearn.svm as svm
import sklearn.metrics as sm
# Organize data sets , Split test set training set
x, y = data.iloc[:, :-1], data['y']
train_x, test_x, train_y, test_y = ms.train_test_split(x, y, test_size=0.25, random_state=7)
model = svm.SVC(kernel='linear')
model.fit(train_x, train_y)
pred_test_y = model.predict(test_x)
print(sm.classification_report(test_y, pred_test_y))
""" precision recall f1-score support 0 0.69 0.90 0.78 40 1 0.83 0.54 0.66 35 accuracy 0.73 75 macro avg 0.76 0.72 0.72 75 weighted avg 0.75 0.73 0.72 75 """
data.head()
""" x1 x2 y 0 5.35 4.48 0 1 6.72 5.37 0 2 3.57 5.25 0 3 4.77 7.65 1 4 2.25 4.07 1 """
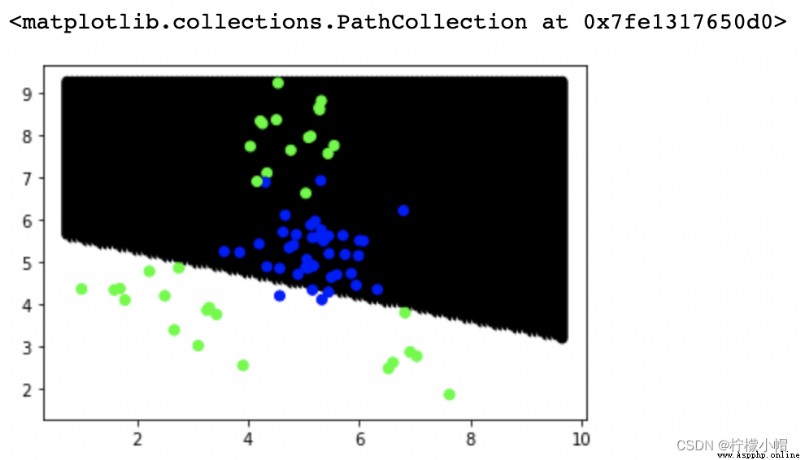
# Violent drawing of classification boundaries
# from x Of min-max, Dismantle 100 individual x coordinate
# from y Of min-max, Dismantle 100 individual y coordinate
# It consists of 10000 Coordinates , Predict the category labels for each coordinate point , Draw a scatter
xs = np.linspace(data['x1'].min(), data['x1'].max(), 100)
ys = np.linspace(data['x2'].min(), data['x2'].max(), 100)
points = []
for x in xs:
for y in ys:
points.append([x, y])
points = np.array(points)
# Predict the category labels for each coordinate point Draw a scatter
point_labels = model.predict(points)
plt.scatter(points[:,0], points[:,1], c=point_labels, cmap='gray')
plt.scatter(test_x['x1'], test_x['x2'], c=test_y, cmap='brg')
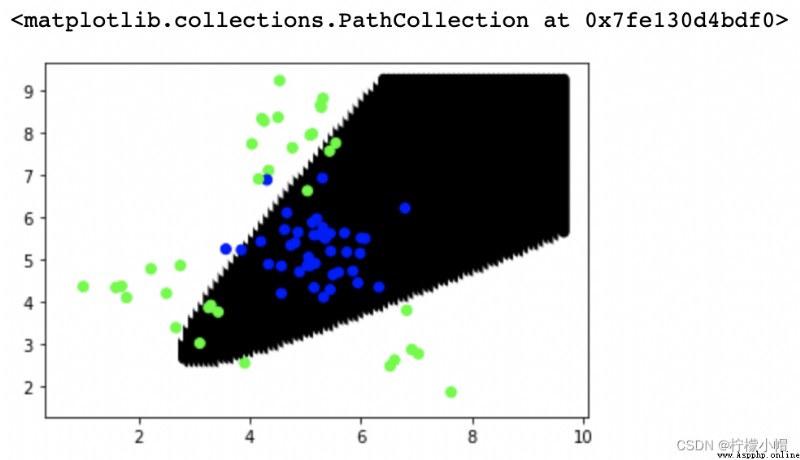
# Polynomial kernel function
model = svm.SVC(kernel='poly', degree=2)
model.fit(train_x, train_y)
pred_test_y = model.predict(test_x)
print(sm.classification_report(test_y, pred_test_y))
# Predict the category labels for each coordinate point Draw a scatter
point_labels = model.predict(points)
plt.scatter(points[:,0], points[:,1], c=point_labels, cmap='gray')
plt.scatter(test_x['x1'], test_x['x2'], c=test_y, cmap='brg')
""" precision recall f1-score support 0 0.84 0.95 0.89 40 1 0.93 0.80 0.86 35 accuracy 0.88 75 macro avg 0.89 0.88 0.88 75 weighted avg 0.89 0.88 0.88 75 """
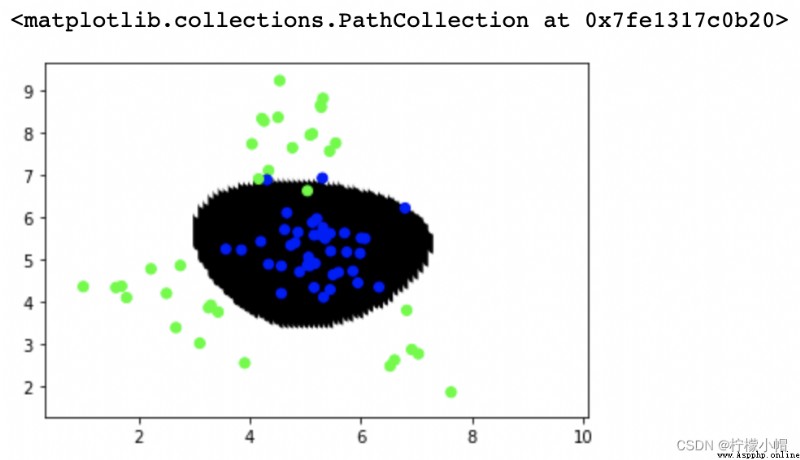
# Radial basis function
model = svm.SVC(kernel='rbf', C=1, gamma=0.1)
model.fit(train_x, train_y)
pred_test_y = model.predict(test_x)
# print(sm.classification_report(test_y, pred_test_y))
# Predict the category labels for each coordinate point Draw a scatter
point_labels = model.predict(points)
plt.scatter(points[:,0], points[:,1], c=point_labels, cmap='gray')
plt.scatter(test_x['x1'], test_x['x2'], c=test_y, cmap='brg')
""" precision recall f1-score support 0 0.97 0.97 0.97 40 1 0.97 0.97 0.97 35 accuracy 0.97 75 macro avg 0.97 0.97 0.97 75 weighted avg 0.97 0.97 0.97 75 """
# The optimal combination of super parameters is found by grid search
model = svm.SVC()
# The grid search
params = [{
'kernel':['linear'], 'C':[1, 10, 100]},
{
'kernel':['poly'], 'degree':[2, 3]},
{
'kernel':['rbf'], 'C':[1, 10, 100], 'gamma':[1, 0.1, 0.001]}]
model = ms.GridSearchCV(model, params, cv=5)
model.fit(train_x, train_y)
pred_test_y = model.predict(test_x)
# print(sm.classification_report(test_y, pred_test_y))
# Predict the category labels for each coordinate point Draw a scatter
point_labels = model.predict(points)
plt.scatter(points[:,0], points[:,1], c=point_labels, cmap='gray')
plt.scatter(test_x['x1'], test_x['x2'], c=test_y, cmap='brg')
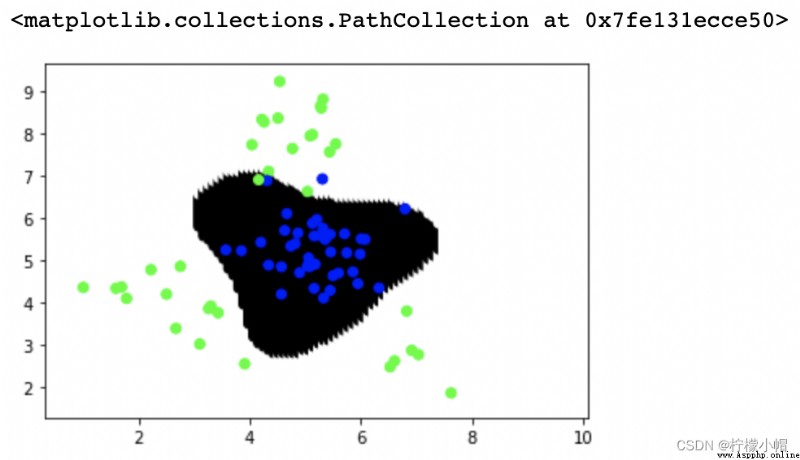
print(model.best_params_)
print(model.best_score_)
print(model.best_estimator_)
""" {'C': 1, 'gamma': 1, 'kernel': 'rbf'} 0.9511111111111111 SVC(C=1, gamma=1) """