This article itself is and 《 Ice lake extraction algorithm based on bimodal threshold segmentation (python Language implementation )》 Together . Random forest is a little more troublesome than threshold segmentation , We need prior knowledge as training data . I also uploaded the training data , You can see : Training data extracted from random forest ice lake _ Random Senli water quality extraction python- Telecommunication document resources -CSDN download
Then there is no need to say more about the algorithm itself , Random forest is also a very mature algorithm , I won't say much about the principle . Let's take a look at the code .
Random forest is a little more complicated , Because a priori data is needed to train the model . But compared with threshold segmentation , Its advantage is that it doesn't have to do DN turn TOA, Mindless training is enough .
def random_forest(img, train_img, train_mask, txt_path):
# Reference resources https://zhuanlan.zhihu.com/p/114069998
######################################## First, train the random forest ##############################################
""" Get training data """
file_write_obj = open(txt_path, 'w')
# Obtain water samples
count = 0
for i in range(train_img.shape[0]):
for j in range(train_img.shape[1]):
# The pixel value of water body category in the label map is 1
if (train_mask[i][j] == 255):
var = ""
for k in range(train_img.shape[-1]):
var = var + str(train_img[i, j, k]) + ","
var = var + "water"
file_write_obj.writelines(var)
file_write_obj.write('\n')
count = count + 1
# Get background samples
Threshold = count
count = 0
for i in range(60000):
X_random = random.randint(0, train_img.shape[0] - 1)
Y_random = random.randint(0, train_img.shape[1] - 1)
# The pixel value of the non water category in the label map is 0
if (train_mask[X_random, Y_random] == 0):
var = ""
for k in range(train_img.shape[-1]):
var = var + str(train_img[X_random, Y_random, k]) + ","
var = var + "non-water"
file_write_obj.writelines(var)
file_write_obj.write('\n')
count = count + 1
if (count == Threshold):
break
file_write_obj.close()
""" Training random forests """
# Read the corresponding txt
from sklearn.ensemble import RandomForestClassifier
from sklearn import model_selection
# Definition dictionary , It is convenient to parse the sample data set txt
def Iris_label(s):
it = {b'water': 1, b'non-water': 0}
return it[s]
path = r"data.txt"
SavePath = r"model.pickle"
# 1. Reading data sets
data = np.loadtxt(path, dtype=float, delimiter=',', converters={7: Iris_label})
# 2. Divide data and labels
x, y = np.split(data, indices_or_sections=(7,), axis=1) # x For data ,y Label
x = x[:, 0:7] # Before selection 7 Bands as features
train_data, test_data, train_label, test_label = model_selection.train_test_split(x, y, random_state=1,
train_size=0.9, test_size=0.1)
# 3. use 100 A tree to create a random forest model , Training random forests
classifier = RandomForestClassifier(n_estimators=100,
bootstrap=True,
max_features='sqrt')
classifier.fit(train_data, train_label.ravel()) # ravel The function is stretched to one dimension
# 4. Calculate the accuracy of random forest
print(" Training set :", classifier.score(train_data, train_label))
print(" Test set :", classifier.score(test_data, test_label))
# 5. Save the model
# Open the file in binary mode :
file = open(SavePath, "wb")
# Write model to file :
pickle.dump(classifier, file)
# Finally close the file :
file.close()
""" Model to predict """
RFpath = r"model.pickle"
SavePath = r"save.png"
################################################ Call the saved model
# Open the file by reading binary
file = open(RFpath, "rb")
# Read the model from the file
rf_model = pickle.load(file)
# Close file
file.close()
################################################ Use the read in model to predict
# Adjust the format of the data before testing
data = np.zeros((img.shape[2], img.shape[0] * img.shape[1]))
# print(data[0].shape, img[:, :, 0].flatten().shape)
for i in range(img.shape[2]):
data[i] = img[:, :, i].flatten()
data = data.swapaxes(0, 1)
# Forecast the data with adjusted format
pred = rf_model.predict(data)
# similarly , We adjust the predicted data to the format of our image
pred = pred.reshape(img.shape[0], img.shape[1]) * 255
pred = pred.astype(np.uint8)
# Save to tif
gdal_array.SaveArray(pred, SavePath)Here is an experimental result :
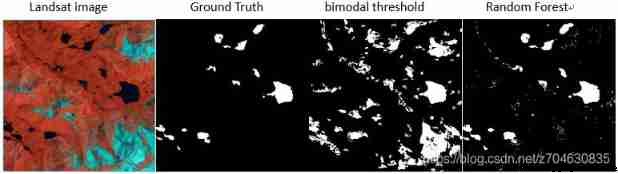
This algorithm was originally compared with the bimodal threshold segmentation algorithm ,( In the absence of any auxiliary data , such as DEM), You can see , Your random forest is much better than bimodal threshold segmentation , At least most of the glaciers can be filtered out . The bimodal threshold segmentation is limited by the choice of threshold , If the threshold is not selected well , Then the glacier will be extracted . When doing large-scale ice lake extraction , It is difficult to use a uniform threshold to extract ice lake , At this time, the bimodal threshold segmentation algorithm The disadvantages will be magnified . Random forests are relatively better , It can avoid the influence of glaciers , But there is a little bit of a problem , Just some meltwater , The river will also be extracted .